Search Count: 24
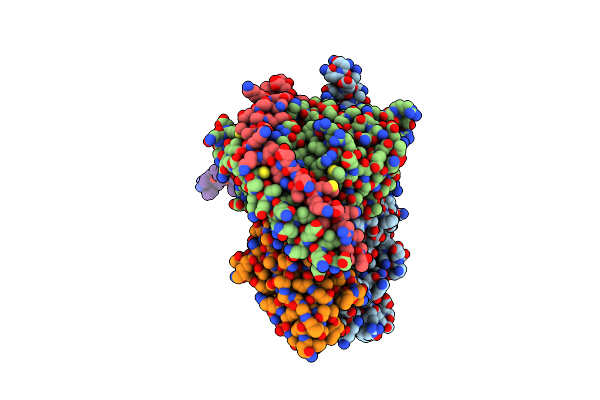 |
Cryo-Em Structure Of Cell-Free Synthesized Human Beta-1 Adrenergic Receptor Cotranslationally Inserted Into A Lipidic Nanodiscs
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: 5FW |
 |
Cryo-Em Structure Of Cell-Free Synthesized Human Histamine H2 Receptor Coupled To Heterotrimeric Gs Protein In Lipid Environment
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: MEMBRANE PROTEIN Ligands: HSM |
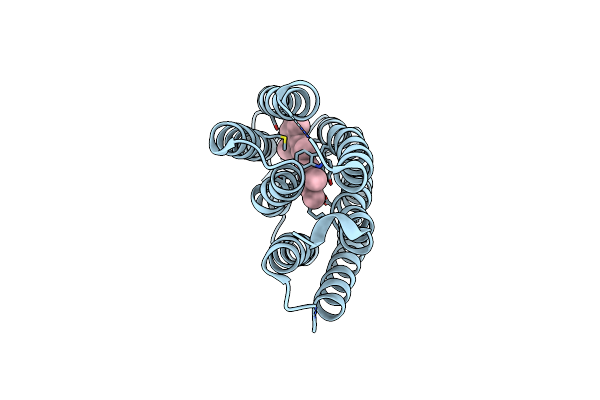 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-14 Classification: ION TRANSPORT Ligands: RET |
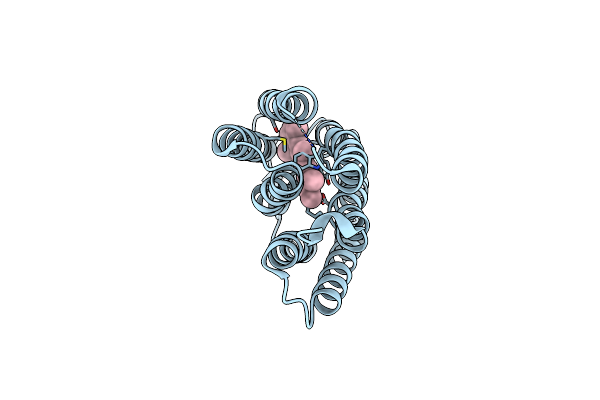 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-09-14 Classification: ION TRANSPORT Ligands: RET |
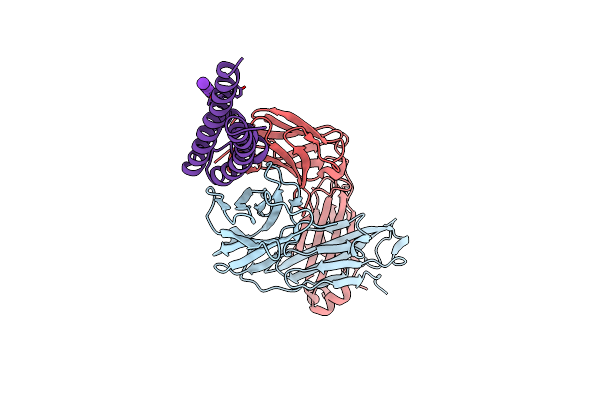 |
Organism: Mus musculus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2016-07-20 Classification: METAL TRANSPORT Ligands: K |
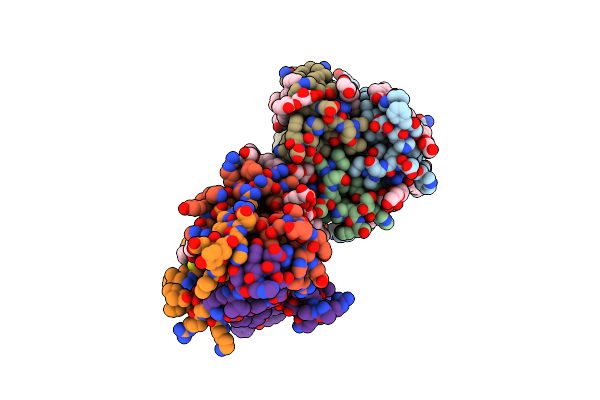 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2014-07-23 Classification: TRANSFERASE Ligands: 78M, 78N, NA, FLC, ZN |
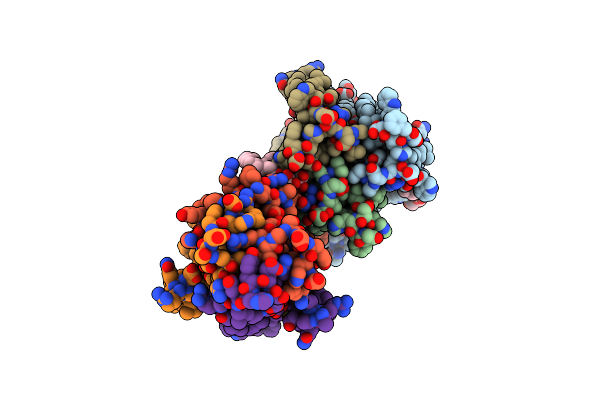 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-05-07 Classification: TRANSFERASE Ligands: 78M, ZN |
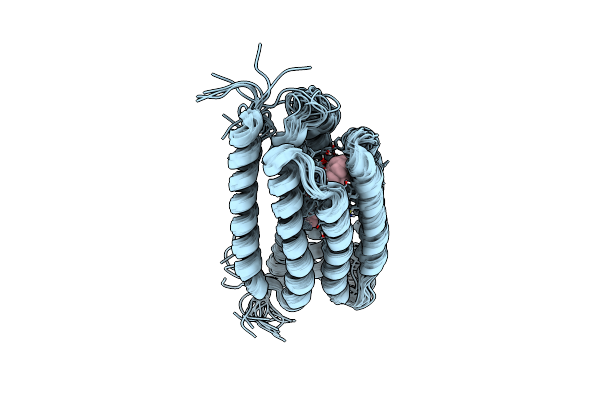 |
Organism: Uncultured marine gamma proteobacterium ebac31a08
Method: SOLUTION NMR Release Date: 2011-11-09 Classification: PROTON TRANSPORT Ligands: RET |
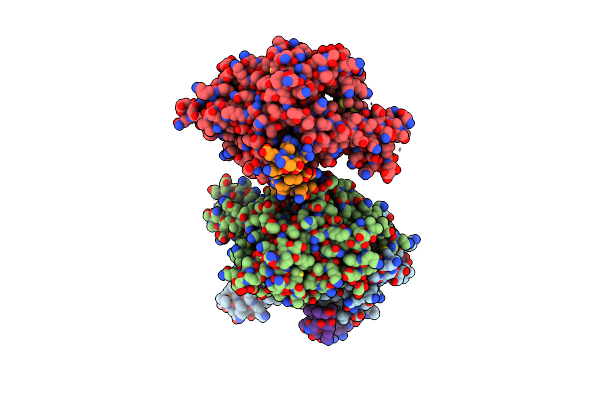 |
Crystal Structure Of S. Cerevisiae Get3 With Bound Adp-Mg2+ In Complex With Get2 Cytosolic Domain
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2011-07-13 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: ADP, MG, ZN |
 |
Crystal Structure Of S. Cerevisiae Get3 In The Open State In Complex With Get1 Cytosolic Domain
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-07-06 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: ZN, PO4 |
 |
Crystal Structure Of S. Cerevisiae Get3 In The Open State In Complex With Get1 Cytosolic Domain
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2011-07-06 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: PO4, ZN |
 |
Crystal Structure Of S.Cerevisiae Get3 In The Semi-Open State In Complex With Get1 Cytosolic Domain
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2011-07-06 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2011-04-06 Classification: PROTEIN BINDING |
 |
Solution Structure Of The E. Coli Outer Membrane Protein Rcsf (Periplasmatic Domain)
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2011-04-06 Classification: SIGNALING PROTEIN |
 |
Complex Structure Of The External Thioesterase Of The Surfactin-Synthetase With A Carrier Domain
Organism: Brevibacillus parabrevis, Bacillus subtilis
Method: SOLUTION NMR Release Date: 2008-12-09 Classification: Ligase/Hydrolase |
 |
 |
Solution Structure Of The E.Coli Rcsc C-Terminus (Residues 700-949) Containing Linker Region And Phosphoreceiver Domain
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2006-09-26 Classification: TRANSFERASE |
 |
Solution Structure Of The E.Coli Rcsc C-Terminus (Residues 700-816) Containing Linker Region
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2006-09-26 Classification: TRANSFERASE |
 |
Solution Structure Of The E.Coli Rcsc C-Terminus (Residues 817-949) Containing Phosphoreceiver Domain
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2006-09-26 Classification: TRANSFERASE |
 |
Organism: Brevibacillus parabrevis
Method: SOLUTION NMR Release Date: 2006-08-01 Classification: LIGASE/TRANSPORT PROTEIN |

