Search Count: 44
 |
Deacetylase Fi8 Utilizes Unconventional Variant Of A Catalytic Triad: The Diluted Triad
Organism: Flavimarina sp. hel_i_48
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: PGE, PEG, EDO, CA |
 |
Crystal Structure Of A Ce20 Carbohydrate Acetylesterase From Pedobacter Psychrotolerans (Ppce20_Ii), With Ancillary Domain
Organism: Pedobacter psychrotolerans
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: PGE, MG, PMS |
 |
Organism: Sphingomonas sp. y57
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: ACT, BGC, ZN, K |
 |
Crystal Structure Of Deacetylase (Hdah) From Vibrio Cholerae In Complex With Saha
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: SHH, ZN, K, ACT |
 |
Crystal Structure Of Dimethoate Hydrolase (Dmha) Of Rhizorhabdus Wittichii In Complex With Octanoic Acid
Organism: Rhizorhabdus wittichii dc-6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: OCA, K, ZN, 1PE |
 |
Crystal Structure Of Rhizorhabdus Wittichii Dimethoate Hydrolase (Dmha) In Complex With Saha
Organism: Rhizorhabdus wittichii dc-6
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: SHH, ZN, K, OCA |
 |
Crystal Structure Of Histone Deacetylase (Hdah) From Vibrio Cholerae In Complex With Decanoic Acid
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: DKA, IMD, ZN, K, NA |
 |
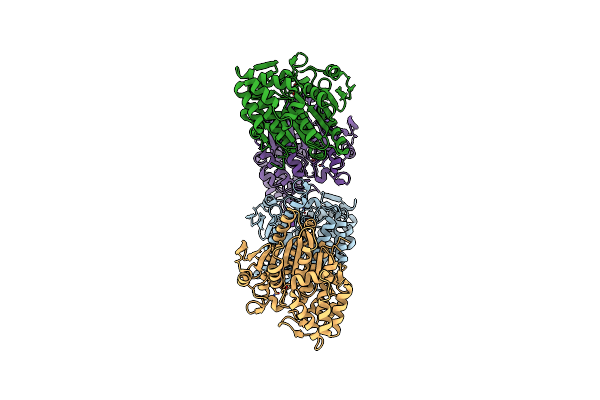
Crystal Structure Of Acetylpolyamine Amidohydrolase (Apah) From Pseudomonas Sp. M30-35
Organism: Pseudomonas sp. m30-35
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: PGE, K, ZN, CL, ACT, PEG |
 |
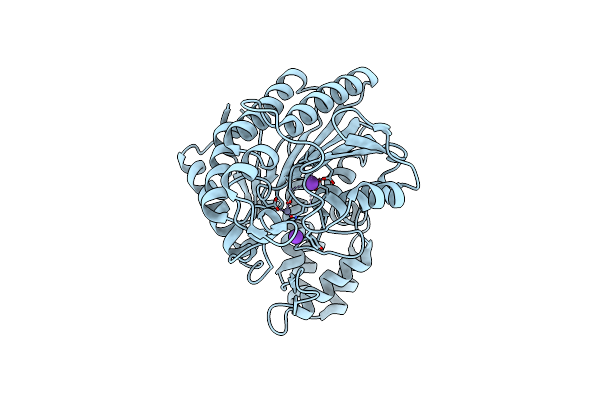
Crystal Structure Of Acetylpolyamine Aminohydrolase (Apah) From Legionella Pneumophila
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: ZN, K |
 |
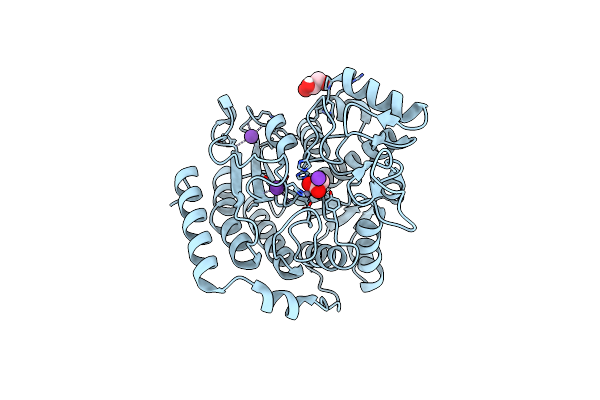
Crystal Structure Of Acetylpolyamine Aminohydrolase (Apah) From Legionella Cherrii
Organism: Legionella cherrii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: ZN, K |
 |
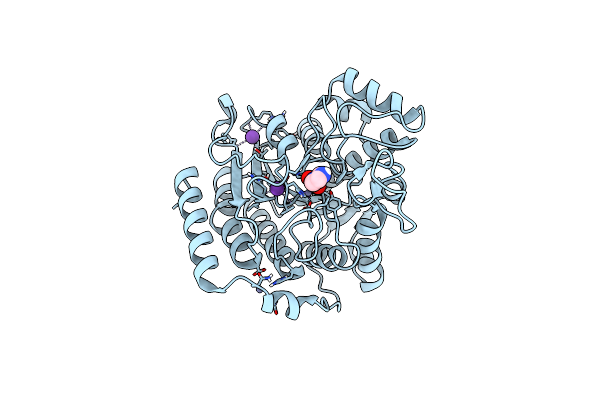
Crystal Structure Of Deacetylase (Hdah) From Klebsiella Pneumoniae Subsp. Ozaenae
Organism: Klebsiella pneumoniae subsp. ozaenae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: GOL, ACT, K, ZN |
 |
Crystal Structure Of Inactive Deacetylase (Hdah) H144A From Klebsiella Pneumoniae Subsp. Ozaenae
Organism: Klebsiella pneumoniae subsp. ozaenae
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: K, ZN, IMD, ACT |
 |
Crystal Structure Of Deacetylase (Hdah) From Klebsiella Pneumoniae Subsp. Ozaenae In Complex With The Inhibitor Saha
Organism: Klebsiella pneumoniae subsp. ozaenae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: K, ZN, SHH |
 |
Crystal Structure Of Deacetylase (Hdah) From Klebsiella Pneumoniae Subsp. Ozaenae In Complex With The Inhibitor Tsa
Organism: Klebsiella pneumoniae subsp. ozaenae
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: K, ZN, PO4, TSN, GOL |
 |
Urethanase Umg-Sp1 Without Inhibitor Or Substrate Displays Flexible Active Site Loops
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-07-24 Classification: HYDROLASE |
 |
Amp-Forming Acetyl-Coa Synthetase From Chloroflexota Bacterium Without Bound Ligand
Organism: Chloroflexota bacterium
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-06-19 Classification: LIGASE Ligands: MG, K |
 |
Amp-Forming Acetyl-Coa Synthetase From Chloroflexota Bacterium With Bound Acetyl Amp
Organism: Chloroflexota bacterium
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-06-19 Classification: LIGASE Ligands: 6R9, EPE, MG, NA |
 |
Organism: Eubacterium ramulus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-11-23 Classification: ISOMERASE Ligands: Q0X, CL, K |
 |
Organism: Eubacterium ramulus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-11-23 Classification: ISOMERASE Ligands: DQH, CL, GOL, SO4, K |
 |
Organism: Eubacterium ramulus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-11-23 Classification: ISOMERASE Ligands: DQH, GOL, SO4, K, CL, EPE |

