Search Count: 35
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: ZN, ADP, PO4, MPO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: ZN, ATP, PO4, MPO |
 |
Crystal Structure Of The Sars-Cov-2 Helicase Nsp13 In Complex With Myricetin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN, PO4, MPO, MYC |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN, PO4, MPO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.70 Å Release Date: 2021-09-08 Classification: SIGNALING PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-08 Classification: DE NOVO PROTEIN Ligands: CA, 38E |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2019-10-16 Classification: SIGNALING PROTEIN Ligands: GTP, MG, 9JE, GDP |
 |
Crystal Structure Of Raga-Q66L-Gtp/Ragc-S75N-Gdp Gtpase Heterodimer Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-10-16 Classification: SIGNALING PROTEIN Ligands: GTP, MG, GDP, 9JE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:5.50 Å Release Date: 2019-10-16 Classification: SIGNALING PROTEIN Ligands: GTP, GDP |
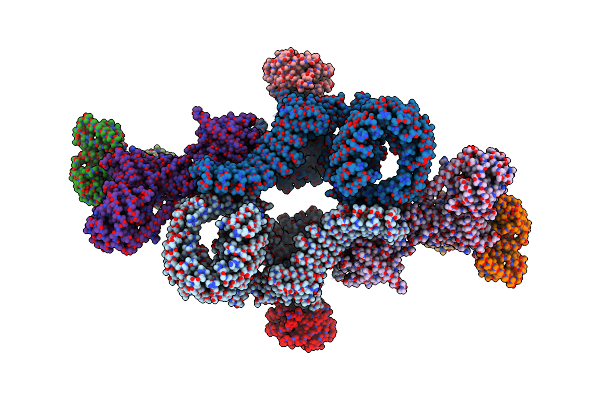 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-10-16 Classification: SIGNALING PROTEIN Ligands: GTP, GDP |
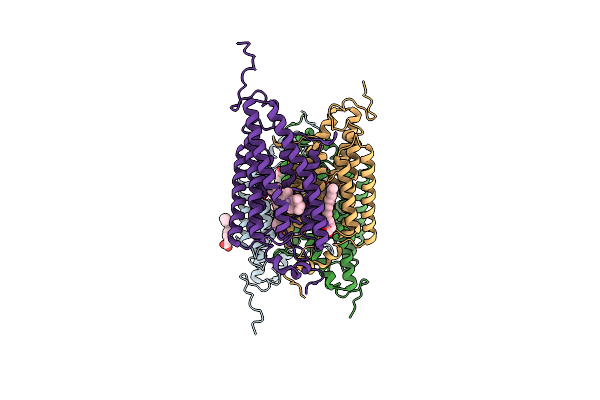 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
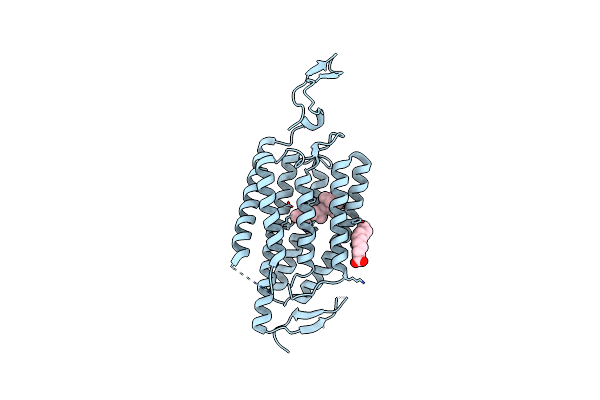 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA, CL |
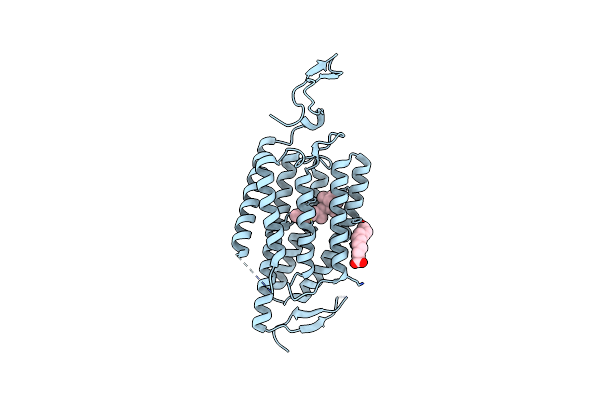 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
 |
X-Ray Diffraction Crystal Structure Of The Murine Pi3K P110Delta In Complex With A Pan Inhibitor
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-09-13 Classification: TRANSFERASE Ligands: 8WH |
 |
Organism: Kluyveromyces marxianus
Method: ELECTRON MICROSCOPY Resolution:6.70 Å Release Date: 2016-04-20 Classification: TRANSFERASE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2013-06-26 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FAD, NH4, NA |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-06-26 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FAD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-06-26 Classification: CIRCADIAN CLOCK PROTEIN |
 |
Crystal Structure Of P110Alpha In Complex With Ish2 Of P85Alpha And The Inhibitor Pik-108
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2011-12-28 Classification: TRANSFERASE Ligands: P08 |
 |
The Crystal Structure Of The Murine Class Ia Pi 3-Kinase P110Delta In Complex With Ic87114.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-02-02 Classification: TRANSFERASE Ligands: IC8 |

