Search Count: 20
 |
Crystal Structure Of Sclam, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellotriosyl-Glucose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellobiosyl-Cellobiose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellotriose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose And A Cellobiose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellohexaose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Laminarihexaose, Presenting A Laminaribiose And A Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: PO4, GLC, CA |
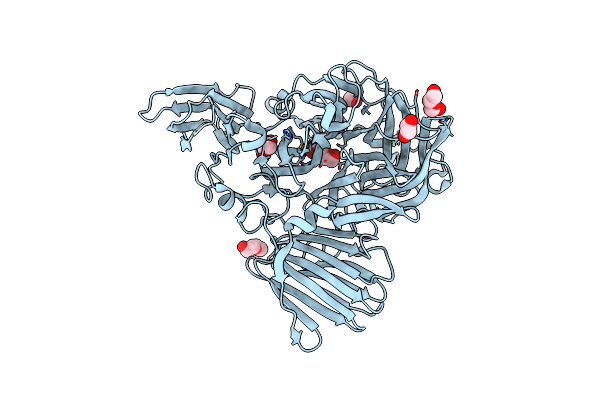 |
Structure Of Glycoside Hydrolase Family 32 From Bifidobacterium Adolescentis
Organism: Bifidobacterium adolescentis (strain atcc 15703 / dsm 20083 / nctc 11814 / e194a)
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2020-01-22 Classification: HYDROLASE Ligands: EDO, PEG |
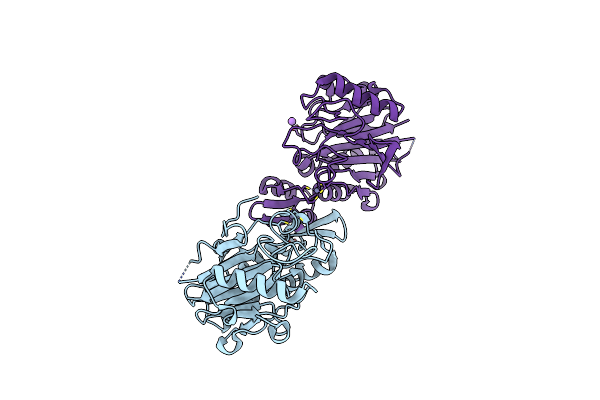 |
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2019-09-11 Classification: UNKNOWN FUNCTION Ligands: ZN, K, CL, NA |
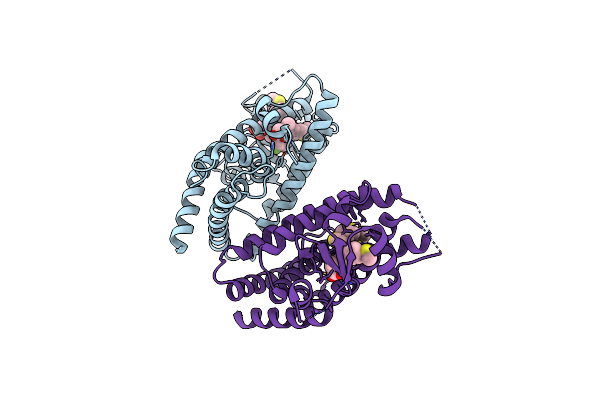 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2016-07-27 Classification: TRANSLATION Ligands: SFI |
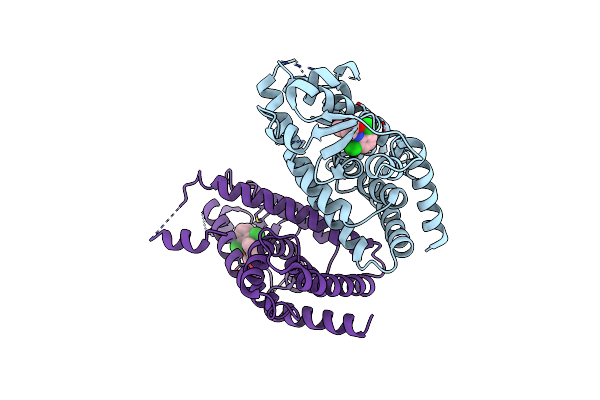 |
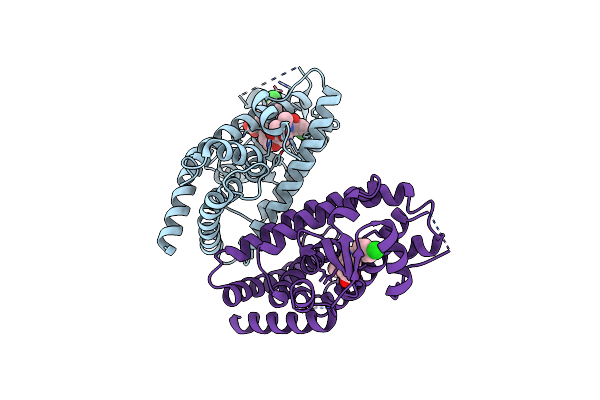
Mechanisms Of Ppargamma Activation By Non-Steroidal Anti-Inflammatory Drugs
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-11-11 Classification: TRANSCRIPTION Ligands: DIF |
 |
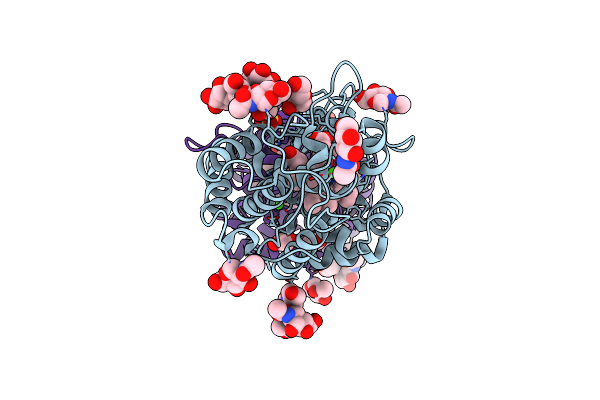
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-11-04 Classification: TRANSLATION Ligands: IMN |
 |
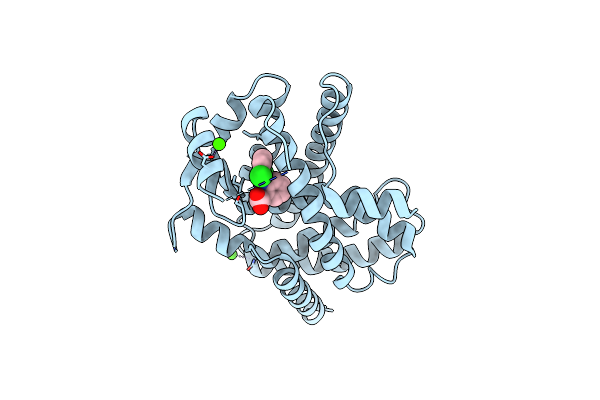
Organism: Trachycarpus fortunei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-05-27 Classification: OXIDOREDUCTASE Ligands: CA, MAN, NAG, SO4, PEG, HEM, EDO, PEO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-04 Classification: TRANSCRIPTION Ligands: DIF, CA |
 |
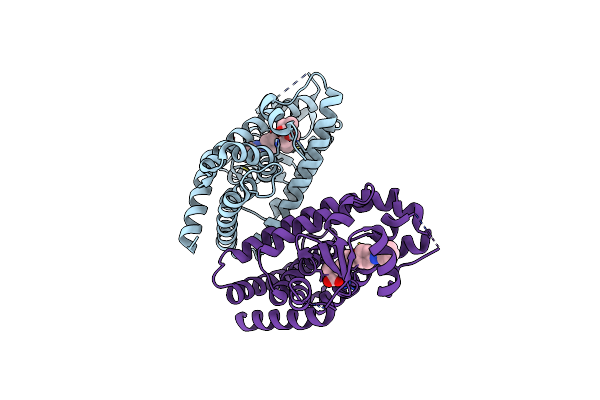
Structural Basis For Gl479 A Dual Peroxisome Proliferator-Activated Receptor Alpha Agonist
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-24 Classification: TRANSFERASE Ligands: Y1N |
 |
Structural Basis For Gl479 A Dual Peroxisome Proliferator-Activated Receptor Gamma Agonist
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-12-24 Classification: NUCLEAR PROTEIN Ligands: Y1N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-05-29 Classification: TRANSCRIPTION Ligands: EDO, WY1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-07-04 Classification: TRANSCRIPTION/TRANSCRIPTION ACTIVATOR Ligands: GW0, GOL |
 |
Human Ppar Gamma Ligand Binding Domain In Complex With Luteolin And Myristic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-03-21 Classification: TRANSCRIPTION Ligands: KNA, LU2, MYR |
 |
Organism: Leptospira interrogans
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2008-03-25 Classification: OXIDOREDUCTASE Ligands: SO4, ZN, FAD |
 |
Organism: Leptospira interrogans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-03-25 Classification: OXIDOREDUCTASE Ligands: ZN, SO4, FAD, NAP |

