Search Count: 24
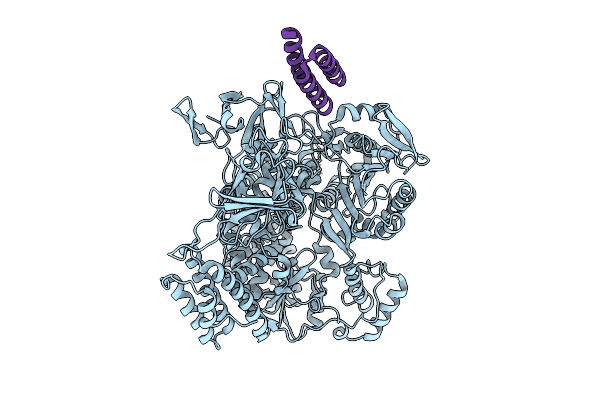 |
De Novo Designed Minibinder Complexed With Clostridioides Difficile Toxin B
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: TOXIN |
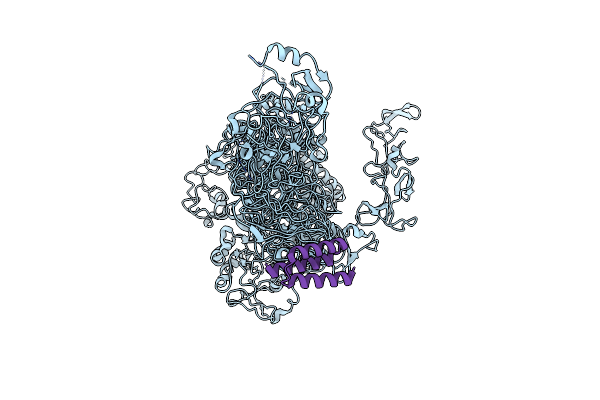 |
Organism: Synthetic construct, Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TOXIN |
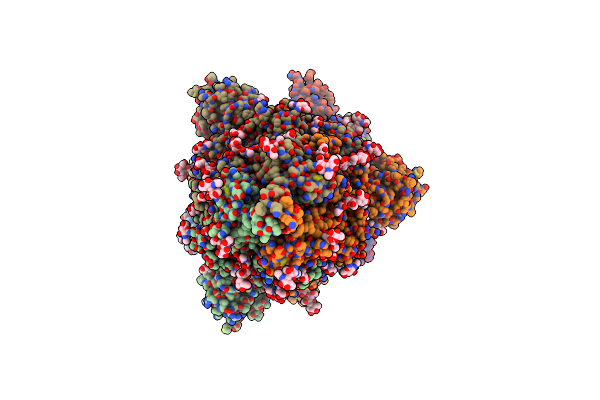 |
Cryoem Structure Of Neutralizing Nanobody Nb30 In Complex With Sars-Cov2 Spike
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-06-16 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
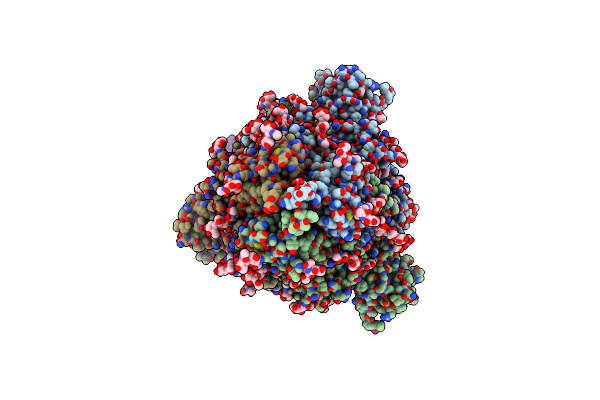 |
Cryoem Structure Of Neutralizing Nanobody Nb12 In Complex With Sars-Cov2 Spike
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-06-16 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-21 Classification: LIPID BINDING PROTEIN |
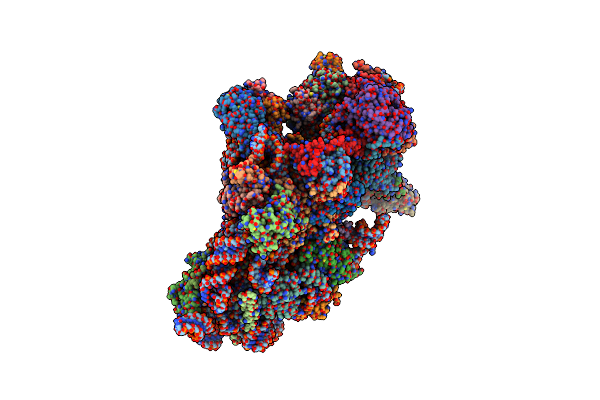 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-10-14 Classification: RIBOSOME/VIRAL PROTEIN Ligands: MG, ZN |
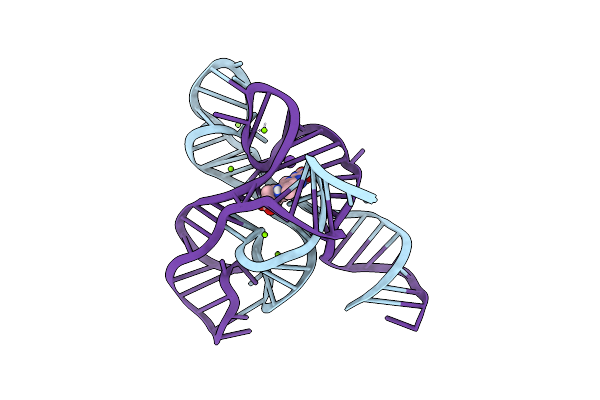 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-09-05 Classification: RNA Ligands: MG, GZ7, K |
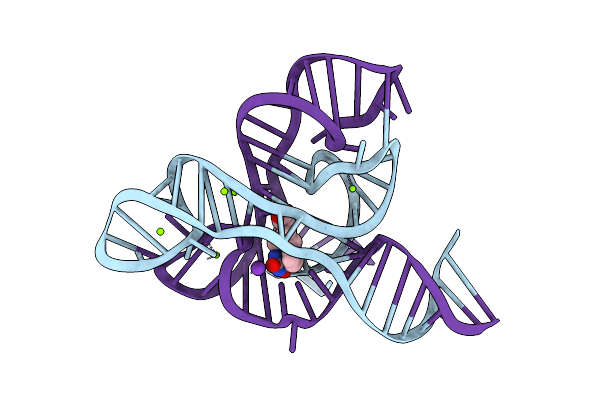 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2018-09-05 Classification: RNA Ligands: MG, GZG, K |
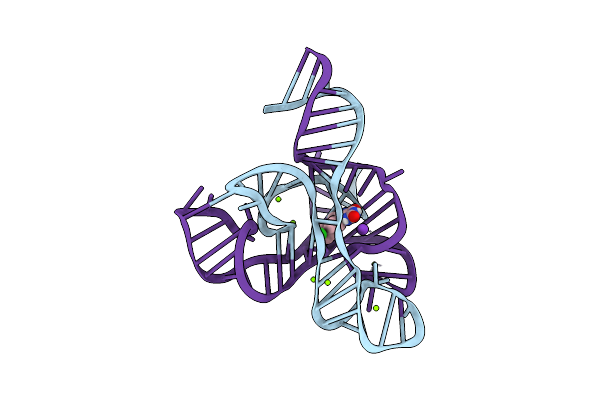 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-05 Classification: RNA Ligands: CL, MG, GZ4, K |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2018-04-25 Classification: IMMUNE SYSTEM Ligands: CDL |
 |
Sumo2 Bound To Mouse Tdp2 Catalytic Domain With A 5'-Phosphorylated Dna Ternary Complex
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-10-11 Classification: hydrolase/dna Ligands: PO4, EDO, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: CA, ACT |
 |
Organism: Lactobacillus rhamnosus lock908
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-10 Classification: RNA Ligands: CS, PRF, MG |
 |
Organism: Faecalibacterium prausnitzii
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-07-01 Classification: RNA Ligands: PRF |
 |
Structure Of A Class Ii Preq1 Riboswitch Reveals Ligand Recognition By A New Fold
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2013-04-17 Classification: RNA Ligands: PRF, CS, MG |
 |
Crystal Structure Of Staphylococcus Aureus Enoyl-Acp Reductase In Complex With Nadp And Afn-1252
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-09-19 Classification: OXIDOREDUCTASE Ligands: 0WD, 0WE |
 |
Method: X-RAY DIFFRACTION
Resolution:2.64 Å Release Date: 2012-08-15 Classification: RNA Ligands: SO4, NCO |
 |
Method: X-RAY DIFFRACTION
Resolution:2.83 Å Release Date: 2012-08-15 Classification: RNA Ligands: SO4, MG |
 |
Method: X-RAY DIFFRACTION
Resolution:2.84 Å Release Date: 2012-08-08 Classification: RNA Ligands: SO4, NCO |
 |
Organism: Bacillus lentus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-04-18 Classification: HYDROLASE Ligands: SO4, CA |

