Search Count: 151
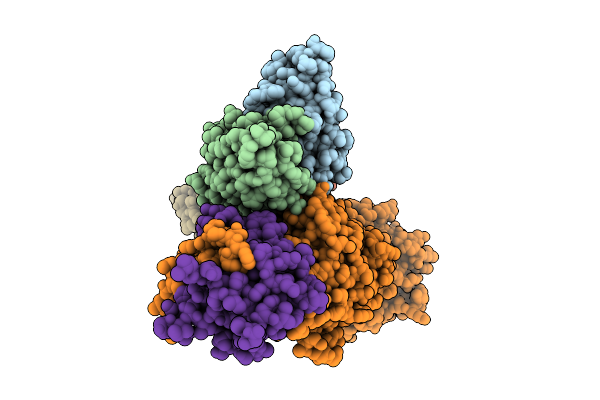 |
Cryo-Em Structure Of Kshv Glycoprotein Ghgl In Complex With Mlkh3 And Mlkh10 Fabs
Organism: Mus musculus, Human gammaherpesvirus 8
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
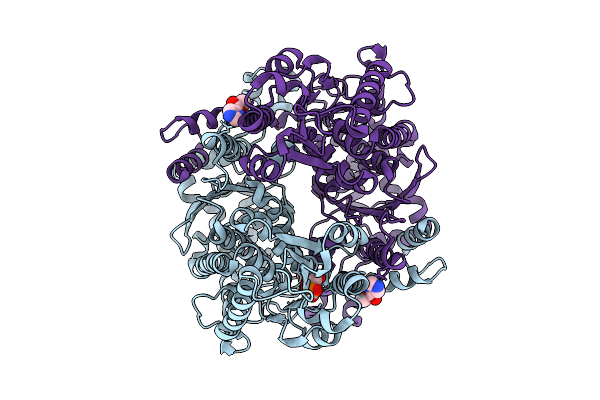
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: PA5, A1IHU |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: A1IIB, PA5, CL |
 |
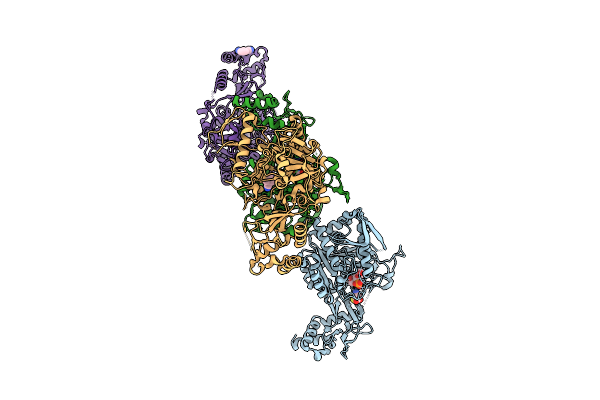
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: MPD, NAD, A1IH7 |
 |
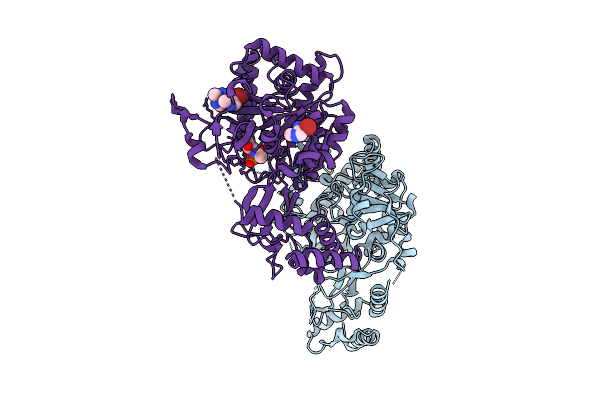
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: GN1, A1IIG, 71G, MG, CL |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: A1IHU, GN1 |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: GN1, YRL, MG |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: YRL, GN1, PO4 |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: A1IIQ, GN1, A1IIG |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With Fragments
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: GN1, A1IHU, A1IIR, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IH9, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerae (Capgi) In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IIA, A1IH8, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment Binder
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IIC |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: APOPTOSIS |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fructose-6-Phosphate
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: F6P, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: A1IHU, PA5 |
 |
Cryoem Structure Of The F Plasmid Relaxosome In Its Pre-Initiation State, Derived From The Ds-27_+143-R Locally-Refined Map 3.76 A
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R Locally-Refined 3.45 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Truncated Trai1-863 In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R_Deltaah+Ctd Locally-Refined 3.42 A Map
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Without Accessory Protein Tram. Derived From The Ss-27_+8Ds+9_+143-R_Deltatram Locally-Refined 2.94 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |

