Search Count: 18
 |
Sc-4E (Scfv Derived From The Mab 4E1 Against Cd93) Crystallized At Ph 7.5 (Dimeric State From Gel Filtration)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: CL, EDO, IMD, NA |
 |
Sc-4E (Scfv Derived From The Mab 4E1 Against Cd93) Crystallized At Ph 7.5 (Monomeric State From Gel Filtration)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: EDO, IMD, NA, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: CL, EDO, IMD, NA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: CL, EDO, IMD, NA |
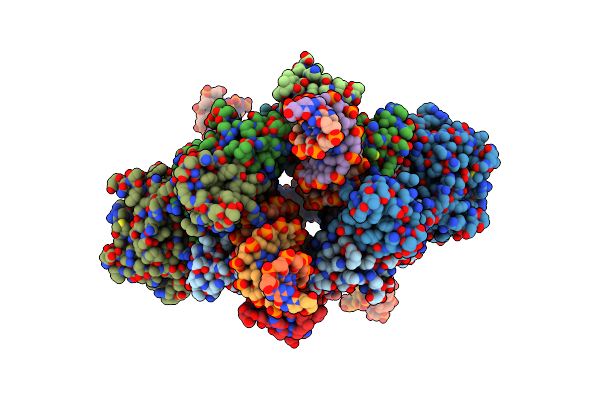 |
Organism: Rous sarcoma virus - prague c
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: VIRAL PROTEIN/DNA Ligands: ZN |
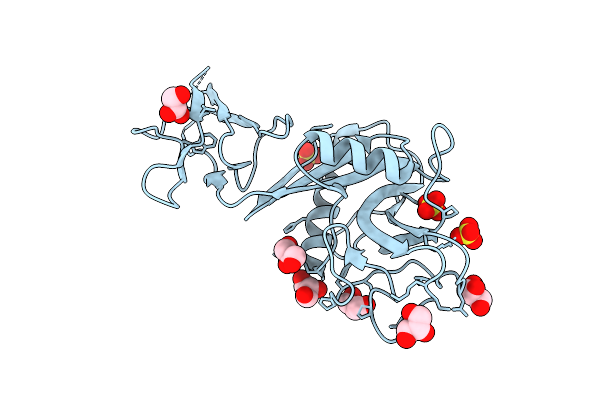 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-11-02 Classification: CELL ADHESION Ligands: GOL, SO4 |
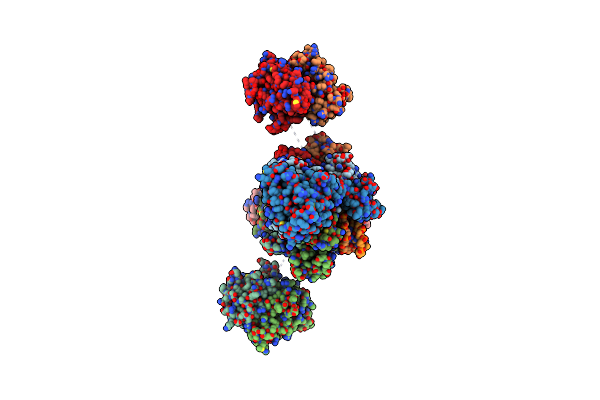 |
Cryo-Em Structure Of Rous Sarcoma Virus Cleaved Synaptic Complex (Csc) With Hiv-1 Integrase Strand Transfer Inhibitor Mk-2048
Organism: Rous sarcoma virus (strain schmidt-ruppin a)
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: HYDROLASE/DNA/INHIBITOR Ligands: ZN, ZZX, MG |
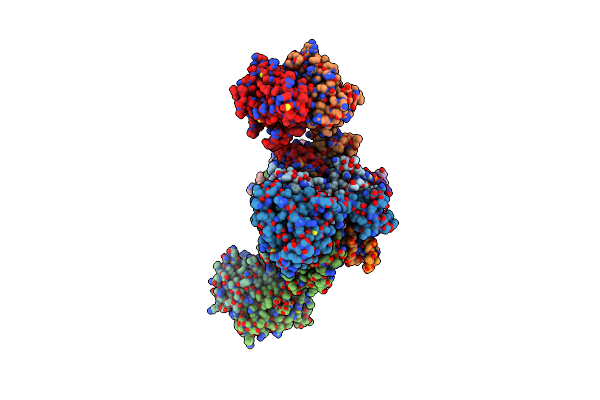 |
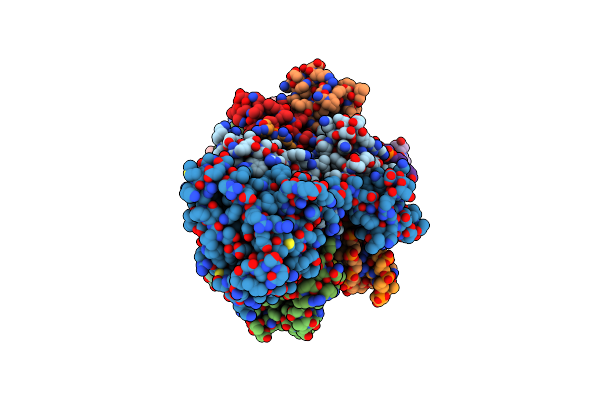
Cryo-Em Structure Of Rous Sarcoma Virus Cleaved Synaptic Complex (Csc) With Hiv-1 Integrase Strand Transfer Inhibitor Mk-2048. Cluster Identified By 3-Dimensional Variability Analysis In Cryosparc.
Organism: Rous sarcoma virus (strain schmidt-ruppin a)
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: HYDROLASE/DNA/INHIBITOR Ligands: ZN, MG, ZZX |
 |
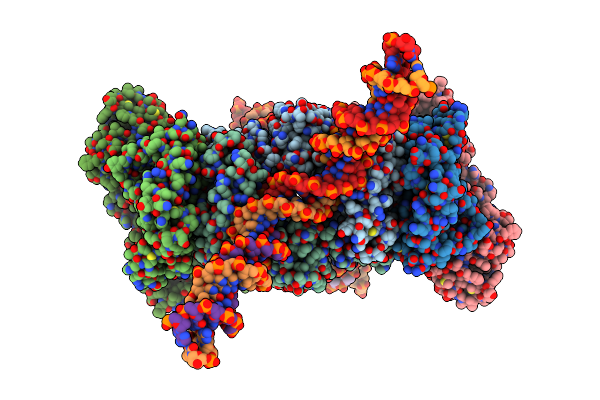
Cryo-Em Structure Of Rous Sarcoma Virus Cleaved Synaptic Complex (Csc) With Hiv-1 Integrase Strand Transfer Inhibitor Mk-2048. Cic Region Of A Cluster Identified By 3-Dimensional Variability Analysis In Cryosparc.
Organism: Rous sarcoma virus (strain schmidt-ruppin a)
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: VIRAL PROTEIN Ligands: ZN, ZZX |
 |
Organism: Saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2), Human t-cell leukemia virus type i, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, MG |
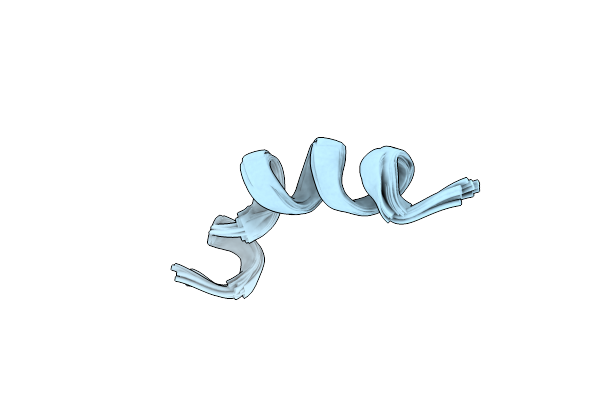 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2017-03-22 Classification: LIPID BINDING PROTEIN |
 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2017-03-22 Classification: LIPID BINDING PROTEIN |
 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2017-03-22 Classification: LIPID BINDING PROTEIN |
 |
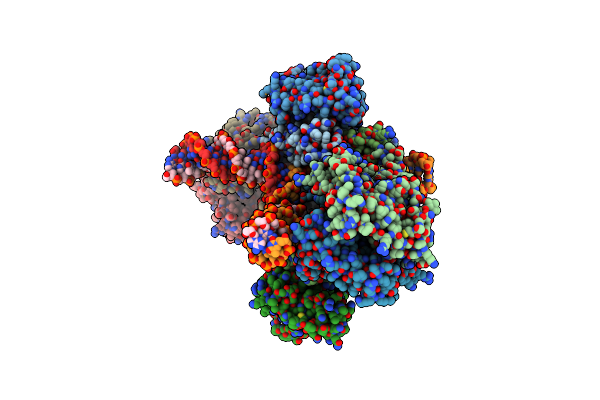
Structural Elucidation Of The Frog Skin-Derived Peptide Esculentin-1A[Esc(1-21)Nh2] Inlipopolysaccharide And Correlation With Their Function
Organism: Pelophylax esculentus
Method: SOLUTION NMR Release Date: 2016-03-02 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Rous sarcoma virus (strain prague c), Rous sarcoma virus (strain schmidt-ruppin e), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2016-02-17 Classification: Transferase/DNA Ligands: ZN, W |
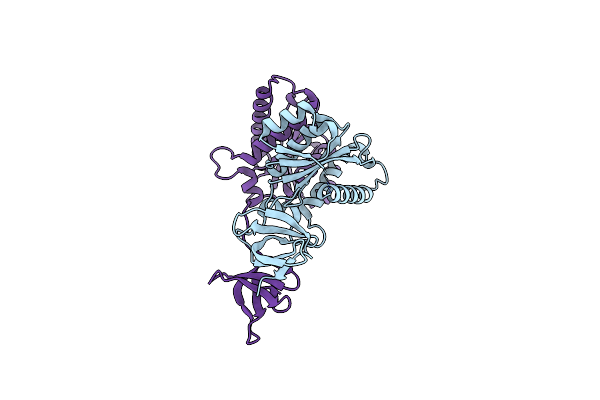 |
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2013-05-15 Classification: DNA BINDING PROTEIN |
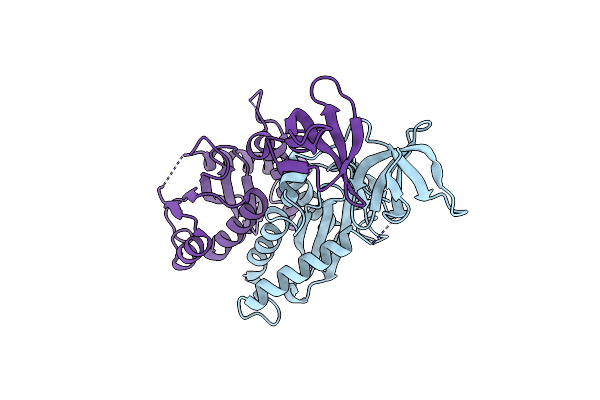 |
Crystal Structure Of Rsv Three-Domain Integrase With Disordered N-Terminal Domain
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-05-15 Classification: DNA BINDING PROTEIN |
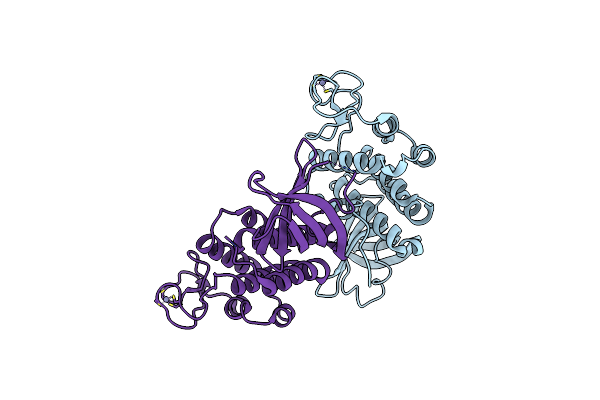 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-10-26 Classification: RECOMBINATION,REPLICATION Ligands: ZN |

