Search Count: 244
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: CO |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Francisella tularensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: APOPTOSIS Ligands: IOD |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct, Acinetobacter baumannii acicu
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: PROTEIN BINDING |
 |
Organism: Synthetic construct, Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: DE NOVO PROTEIN Ligands: PG6, SO4, PG4 |
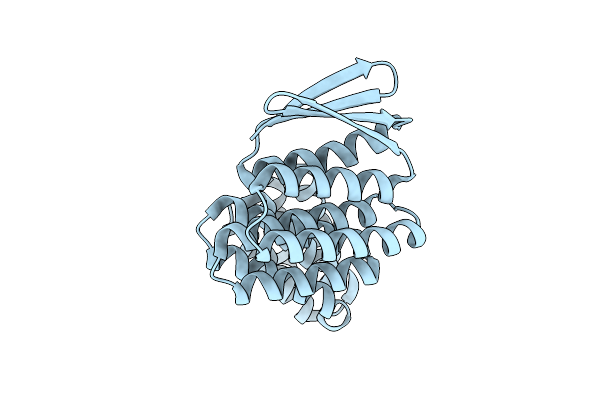 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: CL |
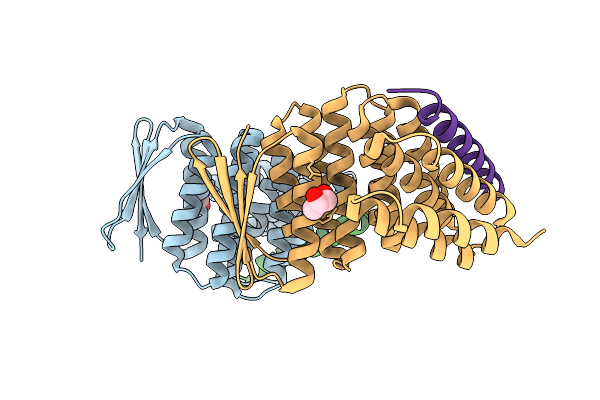 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: PGO |
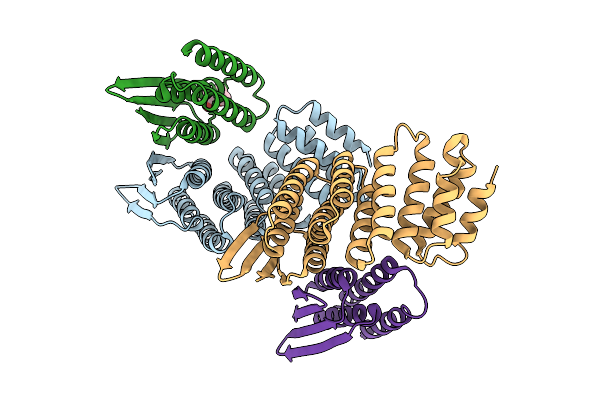 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: MPD |

