Search Count: 17
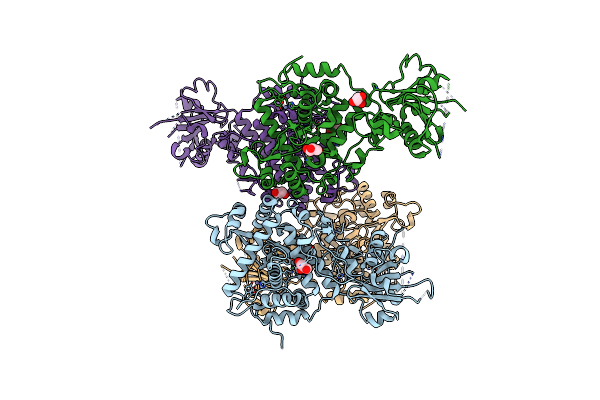 |
Three-Dimensional Structure Of Aip56, A Short-Trip Single Chain Ab Toxin From Photobacterium Damselae Subsp. Piscicida.
Organism: Photobacterium damselae subsp. piscicida
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2023-05-10 Classification: TOXIN Ligands: ZN, NI, GOL |
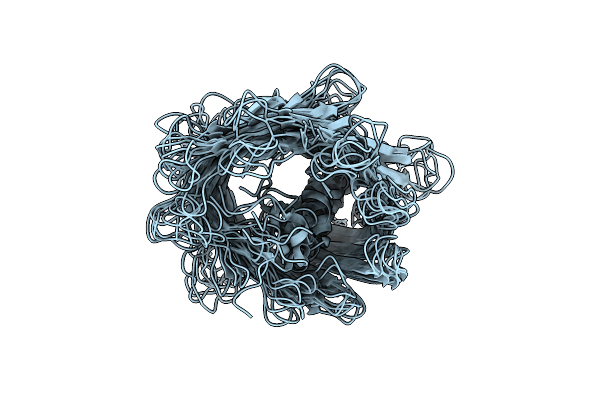 |
Magic-Angle Spinning Nmr Structure Of The Human Voltage-Dependent Anion Channel 1 (E73V/C127A/C232S) In Dmpc Lipid Bilayers
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2022-03-16 Classification: MEMBRANE PROTEIN |
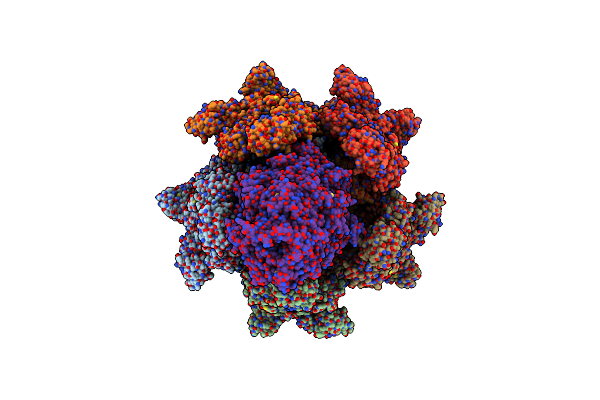 |
Structure Of Photorhabdus Luminescens Tc Holotoxin Pore, Mutation Tccc3-D651A
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2019-11-06 Classification: TOXIN |
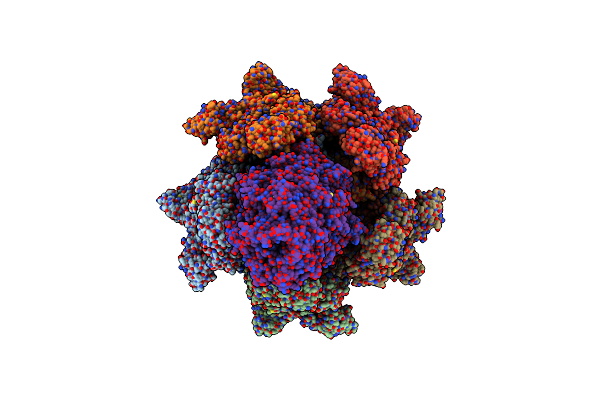 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2019-11-06 Classification: TOXIN |
 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: TOXIN |
 |
Organism: Xenorhabdus nematophila
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: TOXIN |
 |
Organism: Morganella morganii subsp. morganii
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: TOXIN |
 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: TOXIN |
 |
Organism: Yersinia pseudotuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: TOXIN |
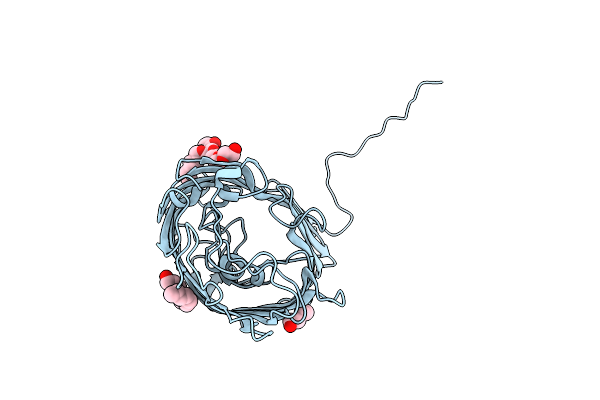 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-10-21 Classification: MEMBRANE PROTEIN Ligands: C8E |
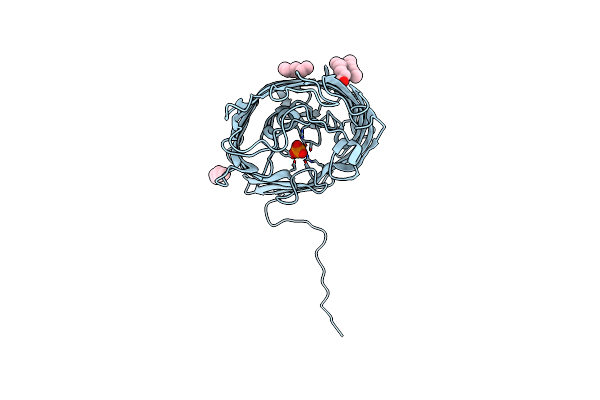 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-10-21 Classification: MEMBRANE PROTEIN Ligands: C8E, PO4 |
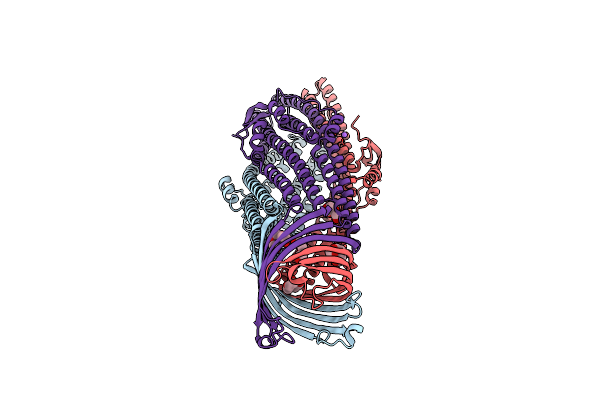 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-01-26 Classification: MEMBRANE PROTEIN Ligands: CL, PGE, NA |
 |
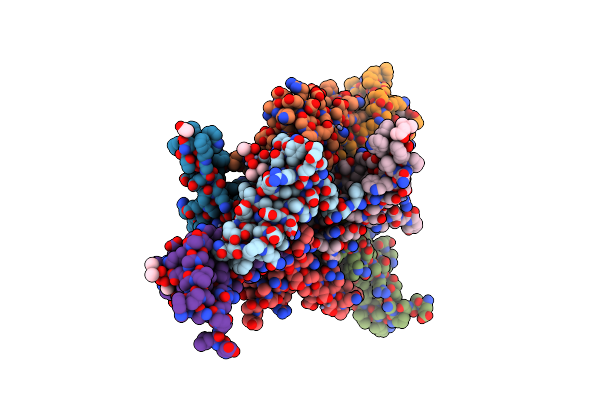
High Resolution Snapshots Of Defined Tolc Open States Present An Iris- Like Movement Of Periplasmic Entrance Helices
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2011-01-26 Classification: TRANSPORT PROTEIN Ligands: CL, LMT |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2008-05-20 Classification: MEMBRANE PROTEIN Ligands: ZN, CAC, ACT, MRD, GVT |
 |
Crystal Structure Of Porb From Corynebacterium Glutamicum (Crystal Form Ii)
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2008-05-20 Classification: MEMBRANE PROTEIN Ligands: ZN, CAC |
 |
Crystal Structure Of Porb From Corynebacterium Glutamicum (Crystal Form Iv)
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2008-05-20 Classification: MEMBRANE PROTEIN Ligands: CAC, ZN |
 |
Crystal Structure Of Porb From Corynebacterium Glutamicum (Crystal Form Iii)
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2008-05-20 Classification: MEMBRANE PROTEIN Ligands: ZN, CAC |

