Search Count: 19
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: CELL CYCLE |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Covalently Inactivated By (S)-But-3-Yn-2-Ylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: A1BC6, FMT |
 |
Proline Utilization A (Puta) From Sinorhizobium Meliloti Inactivated By N-Propargylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: P5F, NAD, SO4, MG, FMT, PEG, PG4, PGE |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With (Prop-2-Ynylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, X79 |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With 2-(Cyanomethylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, X7K, PEG |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With (Allylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, X7Q |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) With The Fad N5 Modified With Propanal Resulting From Inactivation With N-Allylglycine(Replicate #1)
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, FAD, CBG |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) With The Fad N5 Modified With Propanal Resulting From Inactivation With N-Allylglycine (Replicate #2)
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, FAD, CBG, PEG |
 |
Structure Of Proline Utilization A With The Fad Covalently-Modified By Propanal Resulting From Inactivation With N-Allylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, CBG, NAD, MG, SO4, PEG, A1AV8, PGE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-06-19 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-06-19 Classification: TRANSFERASE/PROTEIN BINDING Ligands: ZN |
 |
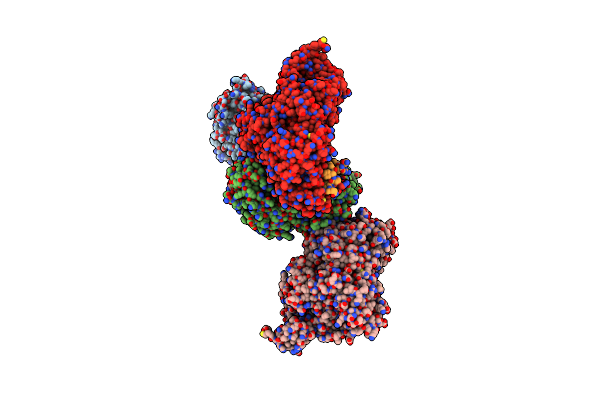
Crystal Structure Of Human O-Glcnac Transferase (Ogt) In Complex With An Exosite-Binding Peptide And Udp-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2023-08-09 Classification: TRANSFERASE Ligands: UD1 |
 |
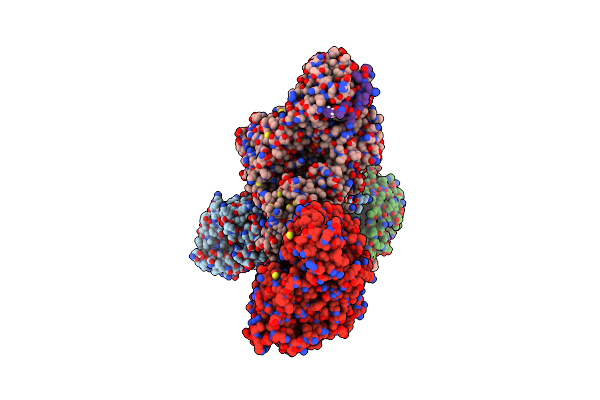
Crystal Structure Of Human O-Glcnac Transferase (Ogt) In Complex With An Exosite-Binding Peptide (Smg9) And Udp-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2023-08-09 Classification: TRANSFERASE Ligands: UD1 |
 |
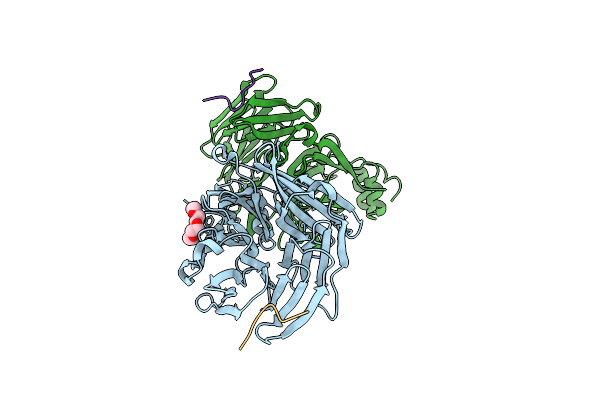
Crystal Structure Of Human O-Glcnac Transferase (Ogt) In Complex With An Exosite-Binding Peptide (Znf831) And Udp-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.69 Å Release Date: 2023-08-09 Classification: TRANSFERASE Ligands: UD1 |
 |
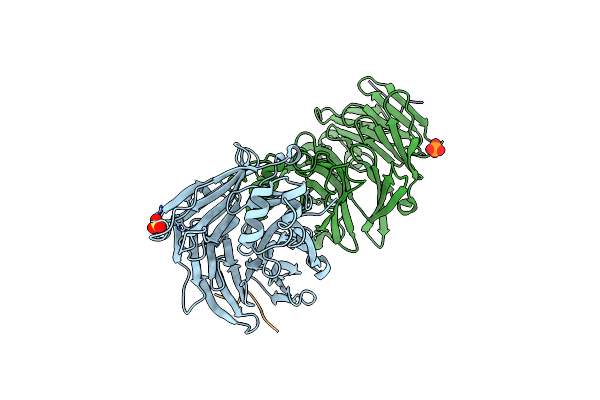
Clathrin Heavy Chain N-Terminal Domain Complexed With Peptide From Protein Mu-Ns Of Reovirus Type 1
Organism: Homo sapiens, Mammalian orthoreovirus 1 lang
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-03-02 Classification: TRANSPORT PROTEIN Ligands: PG4 |
 |
Clathrin Heavy Chain N-Terminal Domain Bound To Non Structured Protein 3 From Eastern Equine Encephalitis Virus
Organism: Homo sapiens, Eastern equine encephalitis virus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-03-02 Classification: TRANSPORT PROTEIN Ligands: SO4, PO4 |
 |
Crystal Structure Of C-Terminal Domain Of Pabpc1 In Complex With Nucleoprotein From Human Coronavirus 229E
Organism: Homo sapiens, Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-03-02 Classification: TRANSCRIPTION Ligands: SO4, GOL |

