Search Count: 111
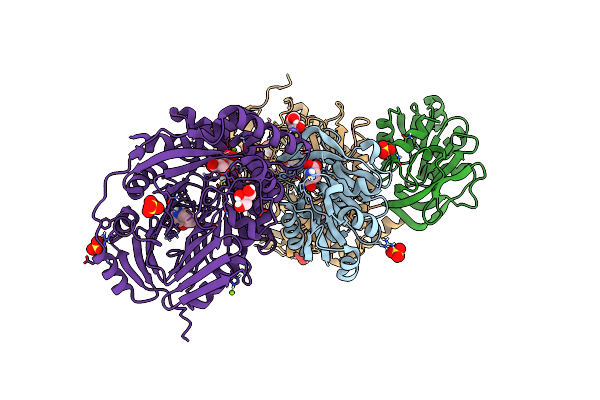 |
Aminodeoxychorismate Synthase Complex From Escherichia Coli, With Glutamine And Chorismate Added
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: GLU, GOL, SO4, ISJ, TRP, MG, CL |
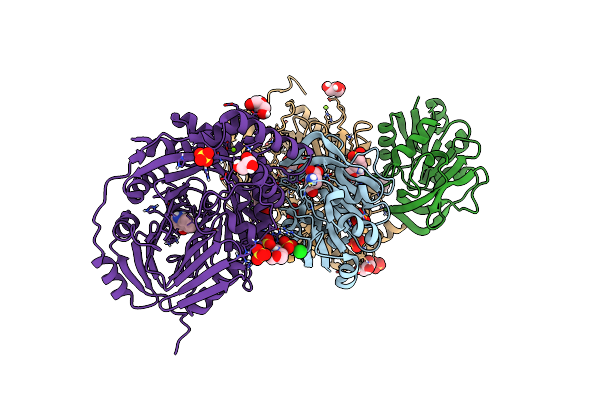 |
Aminodeoxychorismate Synthase Complex From Escherichia Coli, With Glutamine Added
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: GLU, CL, SO4, MES, TRP, MG, GOL |
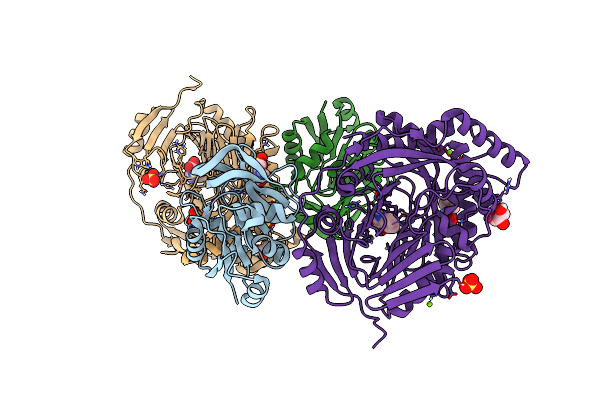 |
Aminodeoxychorismate Synthase Complex From Escherichia Coli, With Edta Added
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: CL, SO4, TRP, MG, MES, GOL |
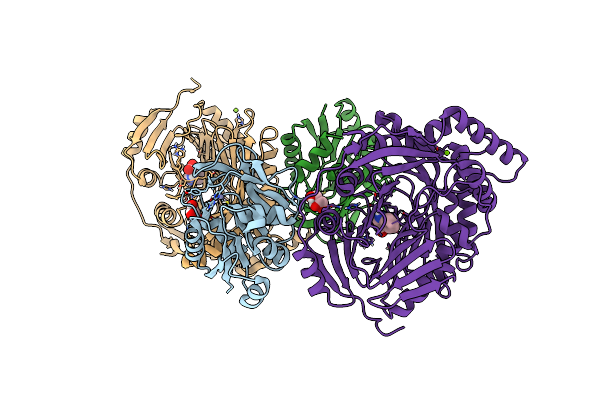 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: ZN, GOL, TRP, MG, CL |
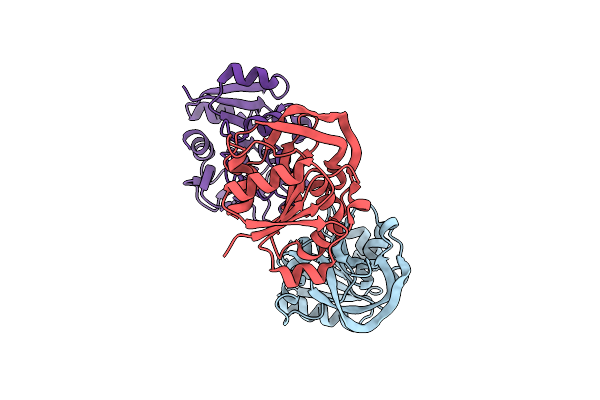 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-01-29 Classification: TRANSFERASE |
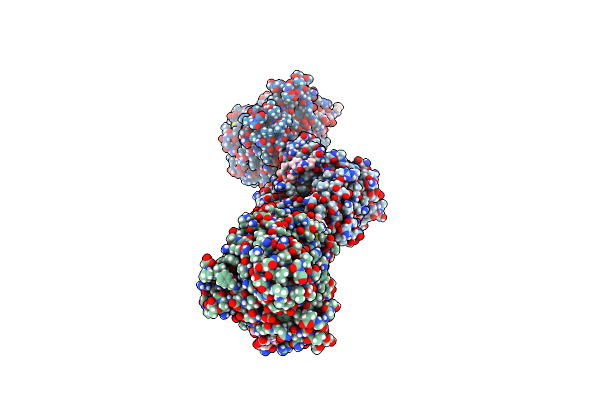 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Epsin-5 Peptide (Lidl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Yap1801 Peptide (Lidm)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS Ligands: GOL, CL |
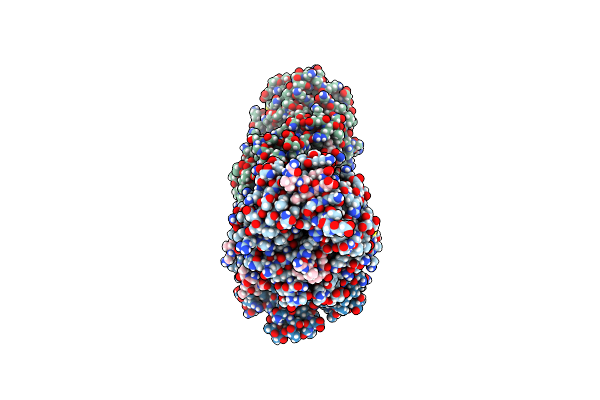 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Epsin-2 Peptide (Lidl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS Ligands: GOL, PO4 |
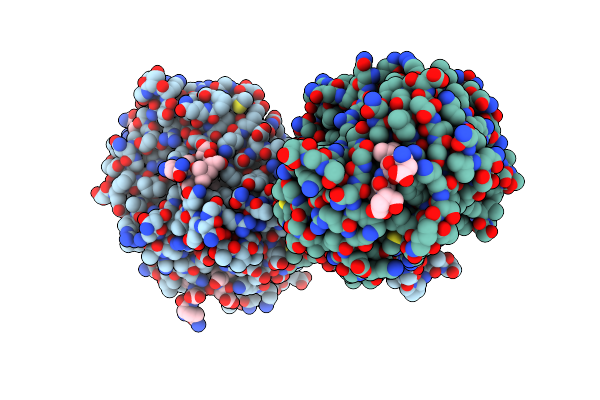 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Apl2/Ap-1 Beta Peptide (Lldl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS |
 |
Crystal Structure Of Yeast Clathrin Heavy Chain N-Terminal Domain Bound To Epsin-1 Peptide (Lidl)
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-11-13 Classification: ENDOCYTOSIS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: A3O, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: DVI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: CIF, GOL |
 |
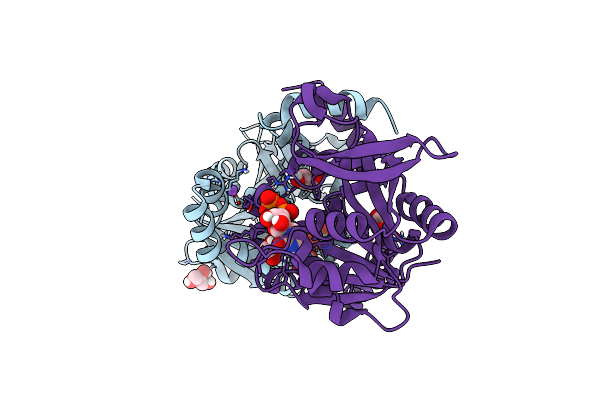
Crystal Structure Of An Hydroxynitrile Lyase Variant (H96A) From Granulicella Tundricola
Organism: Granulicella tundricola
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-10 Classification: LYASE Ligands: 2PE, NM2, MN, PG5, BR, PEG |
 |
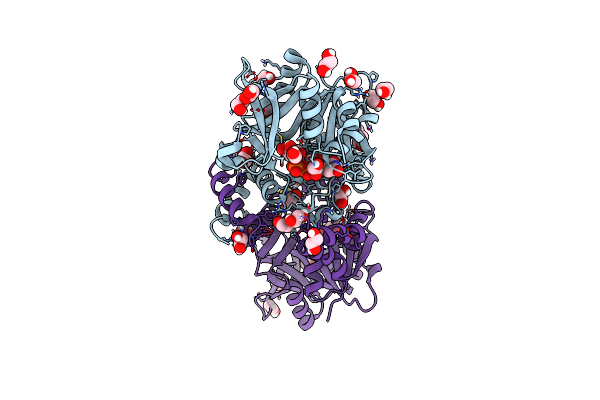
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070 In Complex With Udp
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: UDP, GOL, NA, EDO |
 |
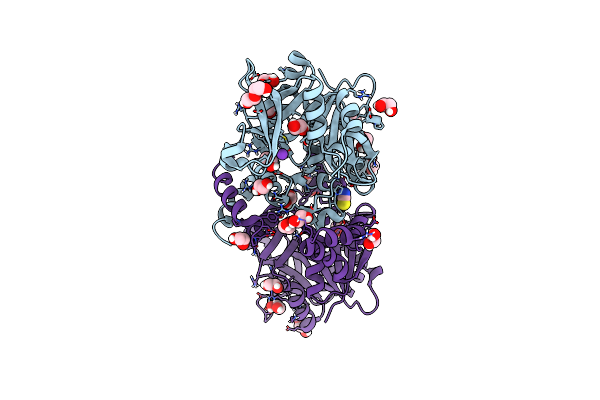
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070 In Complex With Udp-Glucose
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: UPG, EDO, PEG, SCN, MG |
 |
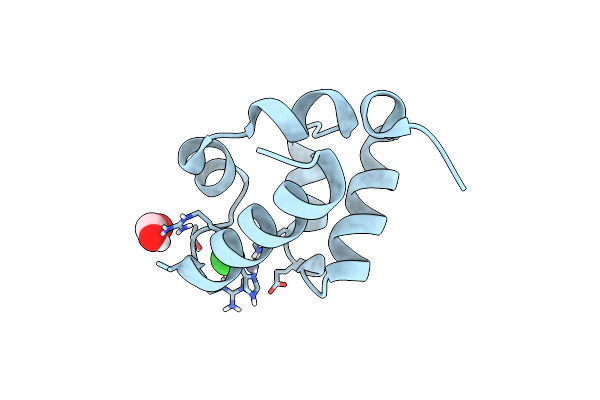
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: PEG, EDO, SCN, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-11-30 Classification: METAL BINDING PROTEIN Ligands: FMT, CL, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-30 Classification: METAL BINDING PROTEIN Ligands: BTB, PEG, CL |
 |
Crystal Structure Of Holo-Swhpa-Mg (Hydroxy Ketone Aldolase) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate And D-Glyceraldehyde
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: LYASE Ligands: 3PY, 3GR, PEG, MG, BR, K |

