Search Count: 34
 |
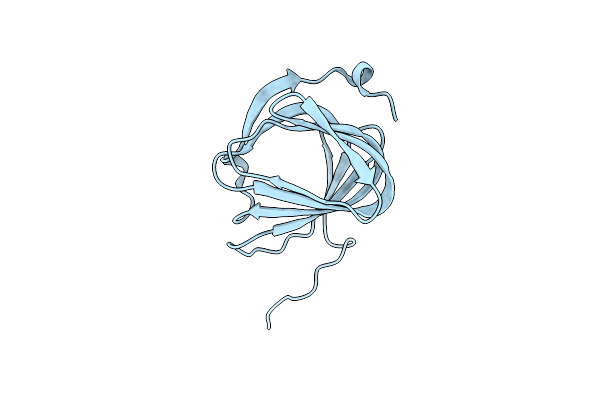
Roll Out The Beta-Barrel: Structure And Mechanism Of Pac13, A Unique Nucleoside Dehydratase
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-03-28 Classification: STRUCTURAL PROTEIN |
 |
Roll Out The Beta-Barrel: Structure And Mechanism Of Pac13, A Unique Nucleoside Dehydratase
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-03-28 Classification: STRUCTURAL PROTEIN |
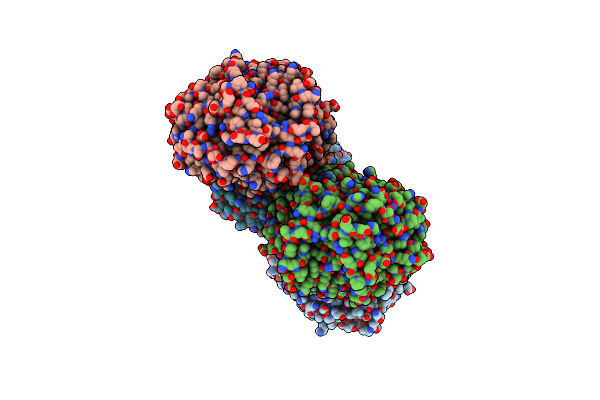 |
Structural Characterization Of The Fast And Promiscuous Macrocyclase From Plant - Pcy1-S562A Bound To Presegetalin F1
Organism: Vaccaria hispanica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-02-07 Classification: HYDROLASE |
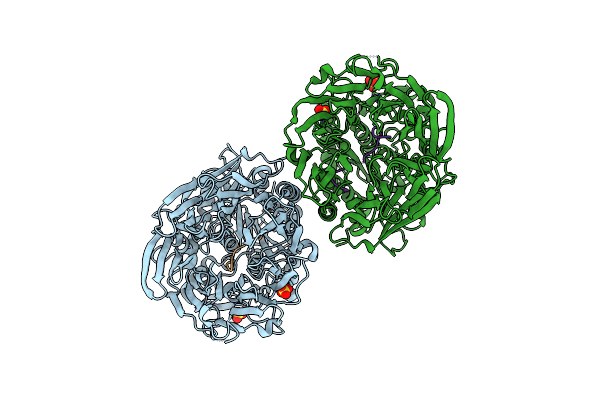 |
Structural Characterization Of The Fast And Promiscuous Macrocyclase From Plant - Pcy1-S562A Bound To Presegetalin B1
Organism: Vaccaria hispanica
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: MG, SO4 |
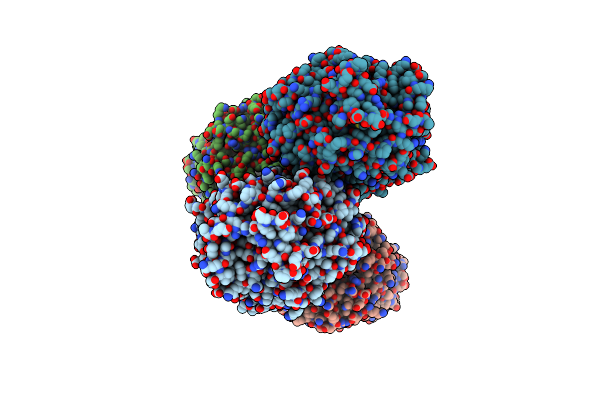 |
Structural Characterization Of The Fast And Promiscuous Macrocyclase From Plant - Pcy1-S562A Bound To Presegetalin A1
Organism: Vaccaria hispanica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: MG, SO4 |
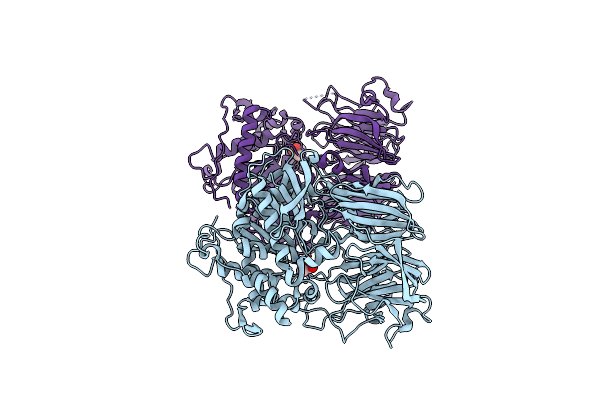 |
Structural Characterization Of The Fast And Promiscuous Macrocyclase From Plant - Apo Pcy1
Organism: Vaccaria hispanica
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: CAC |
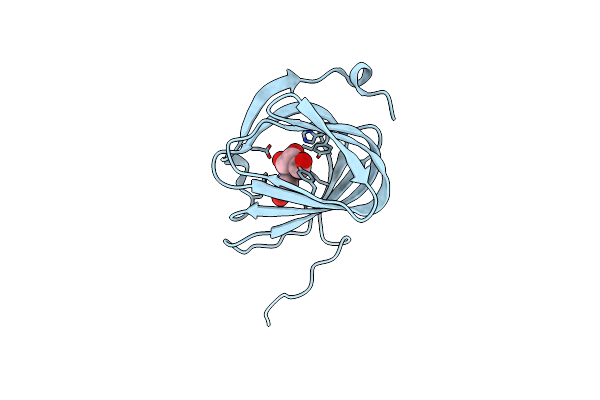 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
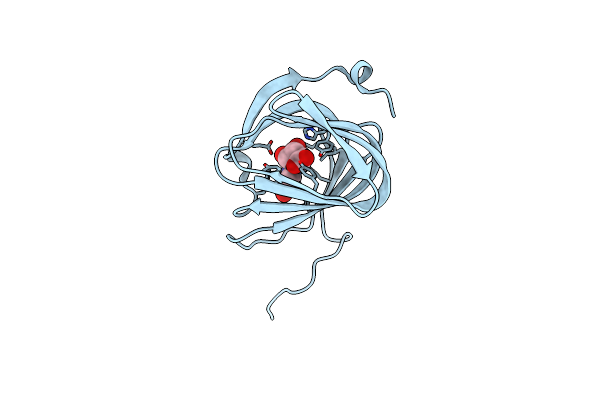 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: UUA |
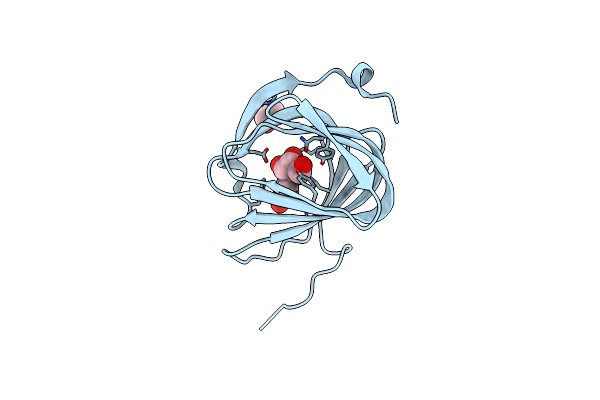 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, URI |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: ZN, 6LK |
 |
The Structure Of Thcox, The First Oxidase Protein From The Cyanobactin Pathways
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141), Cyanothece sp. pcc 7425
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Lyngbya aestuarii, Uncultured prochloron sp.
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: ZN, ADP, MG, PO4 |
 |
Organism: Lyngbya aestuarii, Uncultured prochloron sp.
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: ZN, AMP, CA |
 |
Organism: Lyngbya aestuarii, Uncultured prochloron sp.
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: ZN, ANP, MG |
 |
Organism: Prochloron sp.
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Uncultured prochloron sp. 06037a
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-11-06 Classification: ISOMERASE Ligands: ZN |
 |
Organism: Prochloron didemni
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2013-04-03 Classification: TRANSFERASE |
 |
Crystal Structure Of Syk Kinase Domain With 1-(5-(6,7-Dimethoxyquinolin-4-Yloxy)Pyridin-2-Yl)-3-((1R,2S)-2-Phenylcyclopropyl)Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2012-08-29 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: FPU |

