Search Count: 14
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2023-05-10 Classification: SIGNALING PROTEIN Ligands: ACT, GOL |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2020-12-09 Classification: STRUCTURAL PROTEIN Ligands: CA, GOL, CL |
 |
Organism: Azotobacter vinelandii (strain dj / atcc baa-1303), Salmonella enterica subsp. enterica serovar bovismorbificans
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-12-09 Classification: STRUCTURAL PROTEIN Ligands: ATP, MG |
 |
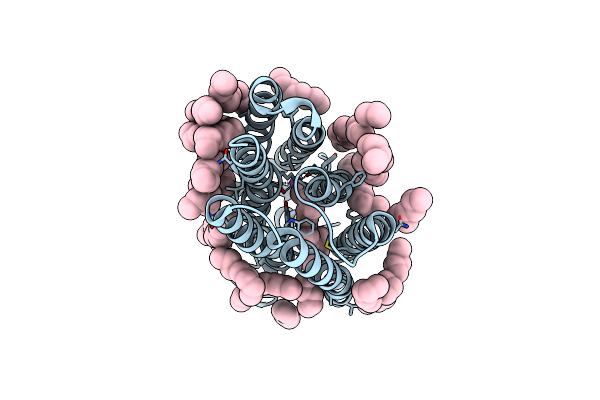
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 20Ms Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, NA |
 |
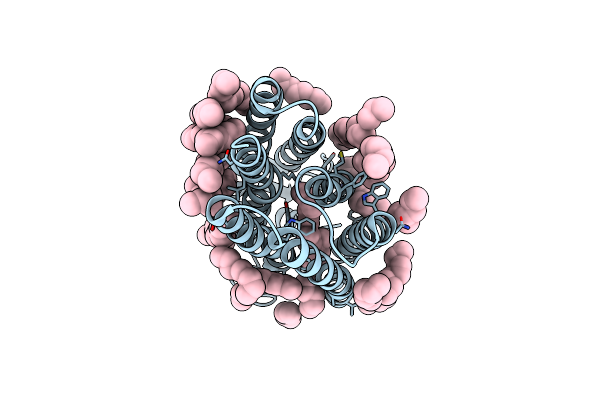
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 1Ms Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, NA |
 |
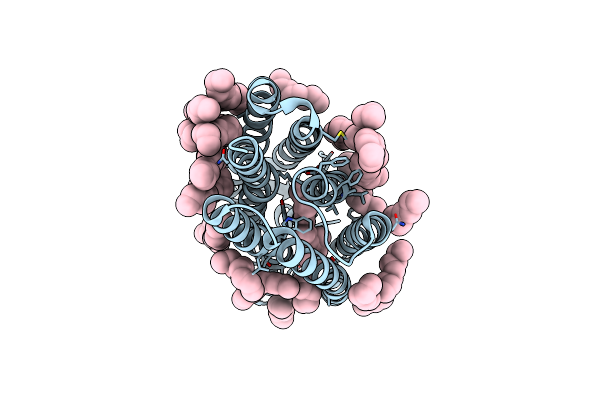
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 30Us+150Us Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 1Ns+16Ns Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: 800Fs+2Ps Structure Of Kr2 With Extrapolated, Light And Dark Datasets
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: Dark Structure In Neutral Conditions With Attached Light Datasets At 800Fs, 2Ps, 100Ps, 1Ns, 16Ns, 1Us, 30Us, 150Us, 1Ms And 20Ms
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Femtosecond To Millisecond Structural Changes In A Light-Driven Sodium Pump: Dark Structure In Acidic Conditions
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Organism: Oryctolagus cuniculus, Bos taurus, Aequorea victoria, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-08 Classification: Fusion protein Ligands: TLA, CA, EDO, GOL, NA |
 |
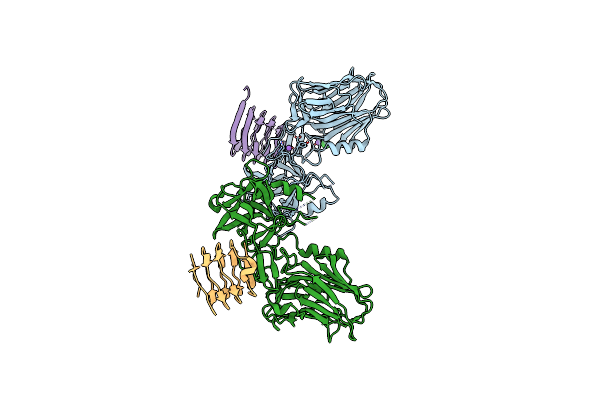
Crystal Structure Of The Bont/A2 Receptor-Binding Domain In Complex With The Luminal Domain Of Its Neuronal Receptor Sv2C
Organism: Clostridium botulinum, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: EDO, 2PE |
 |
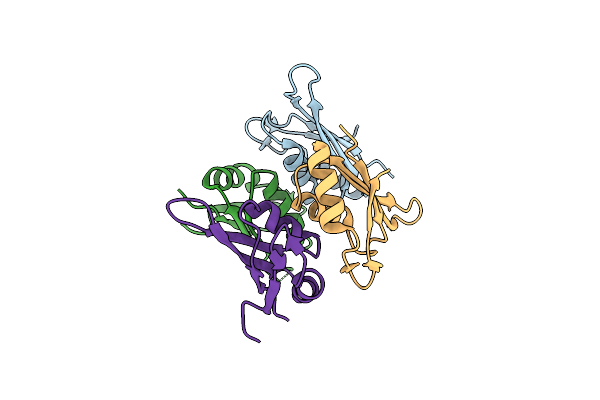
Crystal Structure Of The Botulinum Neurotoxin A Receptor-Binding Domain In Complex With The Luminal Domain Of Sv2C
Organism: Clostridium botulinum, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-11-20 Classification: HYDROLASE Ligands: CL, NA |
 |
The X-Ray Crystal Structure Of The First Rna Recognition Motif (Rrm1) Of The Au-Rich Element (Are) Binding Protein Hur At 2.0 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-03-31 Classification: TRANSCRIPTION |

