Search Count: 51
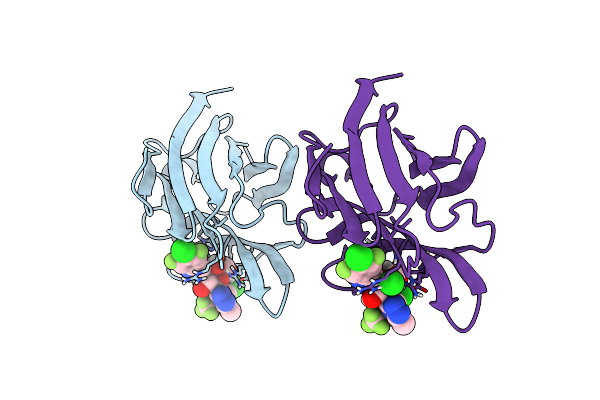 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: A1L0Q, CL |
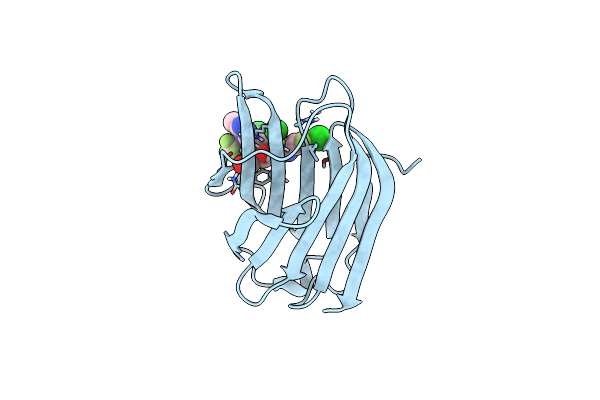 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: A1L0Q, CL |
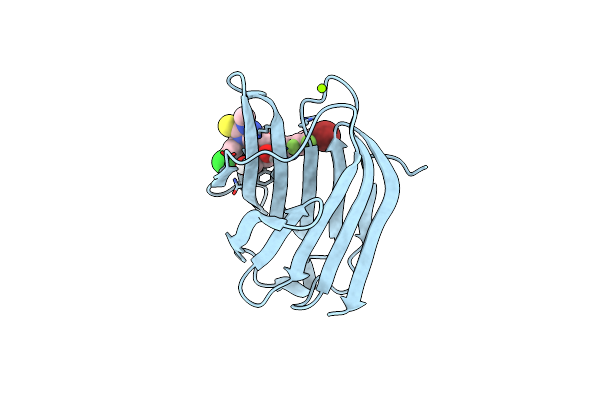 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: MG, A1L0R |
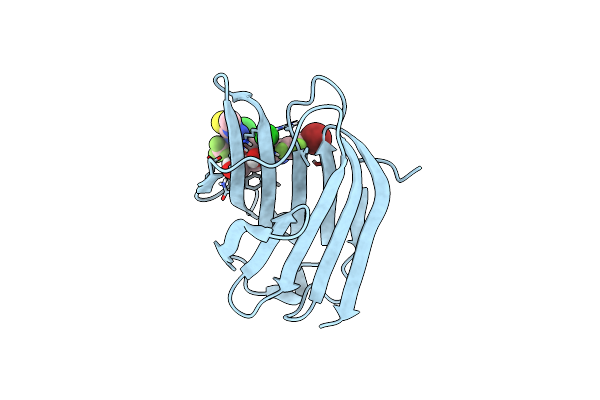 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: A1L0R, CL |
 |
Crystal Structure Of Human Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-03-27 Classification: SUGAR BINDING PROTEIN/INHIBITOR Ligands: CL, SXP, PZE |
 |
Crystal Structure Of Human Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-03-27 Classification: SUGAR BINDING PROTEIN Ligands: CL, QB2, NA |
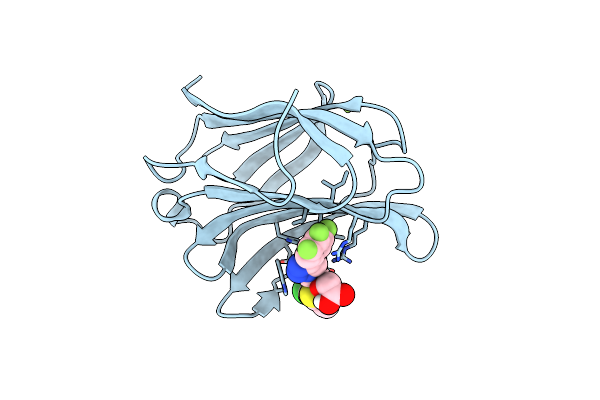 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-03-27 Classification: SUGAR BINDING PROTEIN Ligands: QB2, MG |
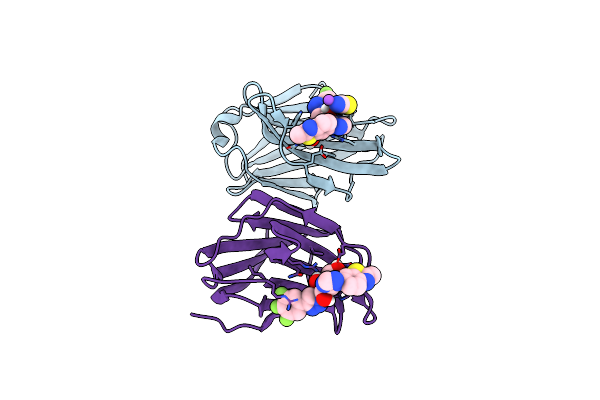 |
Crystal Structure Of Mouse Galectin-3 In Complex With Small Molecule Inhibitor
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: Q1L, SCN, NA |
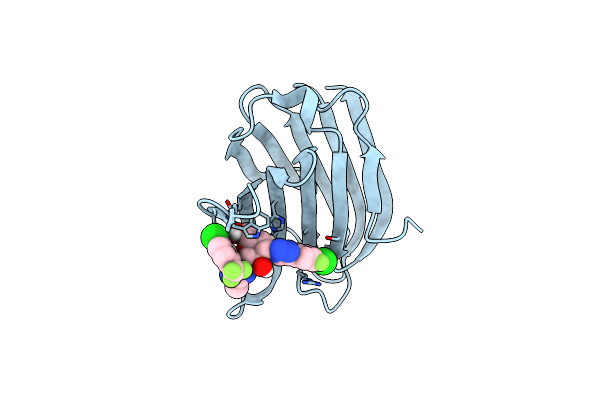 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2022-10-12 Classification: Galactoside-binding lectin Ligands: D9J |
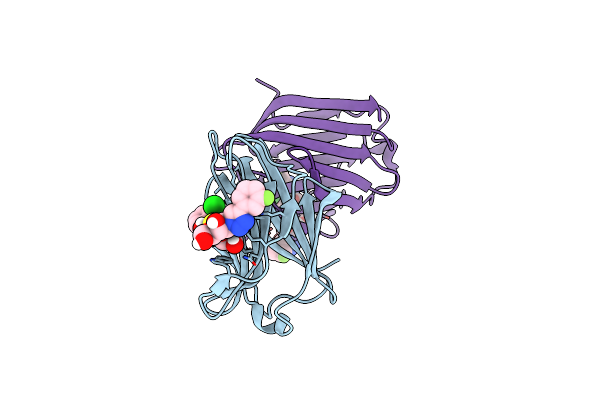 |
Structure Of Human Galectin-3 Crd In Complex With Td-139 Belonging To P31 Space Group.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-09-01 Classification: SUGAR BINDING PROTEIN Ligands: CL, TD2 |
 |
Structure Of Mouse Galectin-3 Crd In Complex With Td-139 Belonging To P6522 Space Group.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2021-09-01 Classification: SUGAR BINDING PROTEIN Ligands: TD2, CL |
 |
Structure Of Mouse Galectin-3 Crd Point Mutant (V160A) In Complex With Td-139 Belonging To P121 Space Group.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-09-01 Classification: SUGAR BINDING PROTEIN Ligands: TD2 |
 |
Xray Structure Of Rat Galectin-3 Crd In Complex With Td-139 Belonging To P121 Space Group
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-09-01 Classification: SUGAR BINDING PROTEIN Ligands: BR, TD2 |
 |
Human Galectin-3 Crd In Complex With Novel Tetrahydropyran-Based Thiodisaccharide Mimic Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2021-06-23 Classification: CARBOHYDRATE Ligands: MG, CL, H5O |
 |
Mouse Galectin-3 Crd In Complex With Novel Tetrahydropyran-Based Thiodisaccharide Mimic Inhibitor
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-23 Classification: CARBOHYDRATE Ligands: H5O |
 |
Crystal Structure Of The Hepatitis C Virus Genotype 2A Strain Jfh1 Ns5B Rna-Dependent Rna Polymerase In Complex With 6-[Ethyl(Methylsulfonyl)Amino]-2-(4-Fluorophenyl)-N-Methyl-5-(3-{[1-(Pyrimidin-2-Yl)Cyclopropyl]Carbamoyl}Phenyl)-1-Benzofuran-3-Carboxamide
Organism: Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2018-11-21 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: J6D, SCN, 2PE, GOL |
 |
Crystal Structure Of The Hepatitis C Virus Genotype 2A Strain Jfh1 L30S Ns5B Rna-Dependent Rna Polymerase In Complex With 6-(Ethylamino)-2-(4-Fluorophenyl)-5-(3-{[1-(5-Fluoropyrimidin-2-Yl)Cyclopropyl]Carbamoyl}-4-Methoxyphenyl)-N-Methyl-1-Benzofuran-3-Carboxamide
Organism: Hepacivirus c
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2018-11-21 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: J6J, SO4, GOL, PG4 |
 |
Crystal Structure Of The Hepatitis C Virus Ns5B Rna-Dependent Rna Polymerase In Complex With 2-(4-Fluorophenyl)-N-Methyl-6-[(Methylsulfonyl)Amino]-5-(Propan-2-Yloxy)-1-Benzofuran-3-Carboxamide
Organism: Hepatitis c virus genotype 1b (isolate con1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-05-10 Classification: TRANSFERASE Ligands: 23E, 2N5, SO4, GOL, PG4 |
 |
Crystal Structure Of The Hepatitis C Virus Ns5B Rna-Dependent Rna Polymerase In Complex With 2-({3-[1-(2-Cyclopropylethyl)-6-Fluoro-4-Hydroxy-2-Oxo-1,2-Dihydroquinolin-3-Yl]-1,1-Dioxo-1,4-Dihydro-1Lambda~6~,2,4-Benzothiadiazin-7-Yl}Oxy)Acetamide
Organism: Hepatitis c virus genotype 1b (isolate con1)
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2017-05-10 Classification: TRANSFERASE Ligands: 23E, 8XV, SO4 |
 |
Crystal Structure Of The Hepatitis C Virus Ns5B Rna-Dependent Rna Polymerase In Complex With 3-[2-(4-Fluorophenyl)-3-(Methylcarbamoyl)-1-Benzofuran-5-Yl]Benzoic Acid
Organism: Hepatitis c virus genotype 1b (isolate con1)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2017-05-10 Classification: TRANSFERASE Ligands: 23E, SO4, 8XS, GOL |

