Search Count: 30
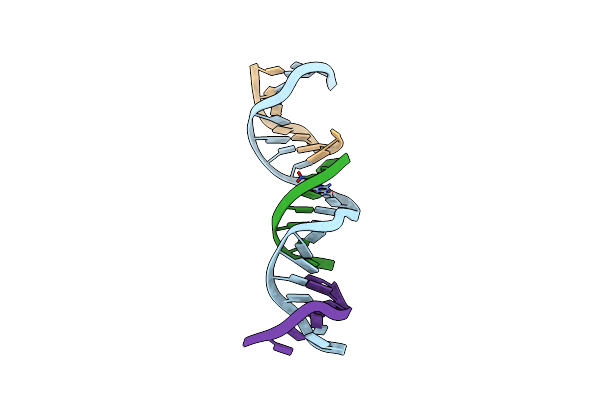 |
[Zp] Self-Assembling Dna Crystal With Expanded Genetic Code Using C,A,T,G, Z And P Nucleotides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2025-01-22 Classification: DNA |
 |
E. Coli Dna-Directed Rna Polymerase Transcription Elongation Complex Bound To The Unnatural Dz-Ptp Base Pair In The Active Site
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: ZN, MG, S9F |
 |
E. Coli Dna-Directed Rna Polymerase Transcription Elongation Complex Bound The Unnatural Ds-Btp Base Pair In The Active Site
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG, X0F |
 |
E. Coli Dna-Directed Rna Polymerase Transcription Elongation Complex Bound The Unnatural Db-Utp Base Pair In The Active Site
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSCRIPTION/DNA/RNA Ligands: DGP, ZN, MG, UTP |
 |
E. Coli Dna-Directed Rna Polymerase Transcription Elongation Complex Bound The Unnatural Db-Stp Base Pair In The Active Site
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSCRIPTION/DNA/RNA Ligands: ZN, MG, X0O |
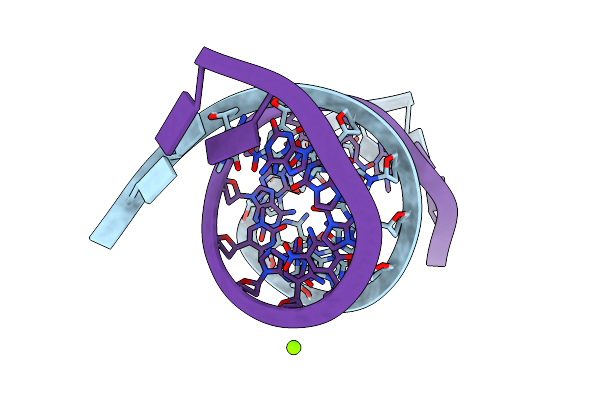 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-05-17 Classification: DNA Ligands: MG |
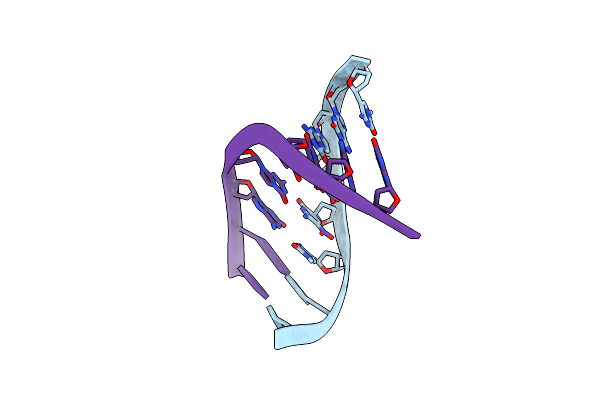 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-05-17 Classification: DNA |
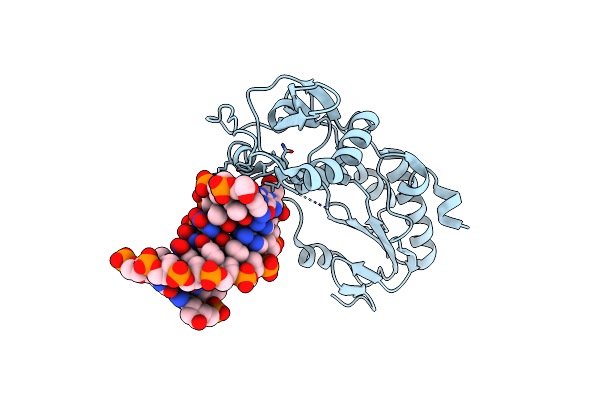 |
Crystal Structure Of Alien Dna Ctszzpbsbszppbag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-17 Classification: DNA BINDING PROTEIN/DNA |
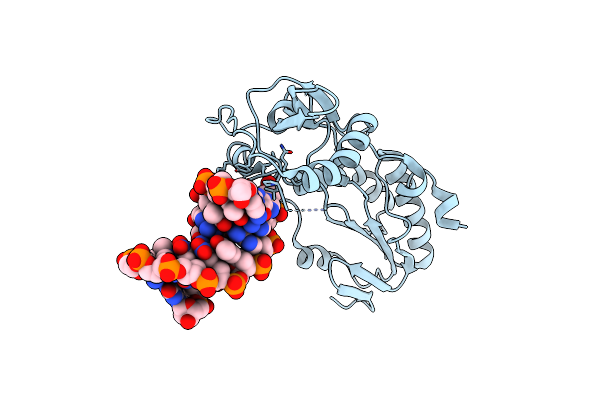 |
Crystal Structure Of B-Form Alien Dna 5'-Cttbppbbsszzsaag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-10 Classification: DNA BINDING PROTEIN/DNA |
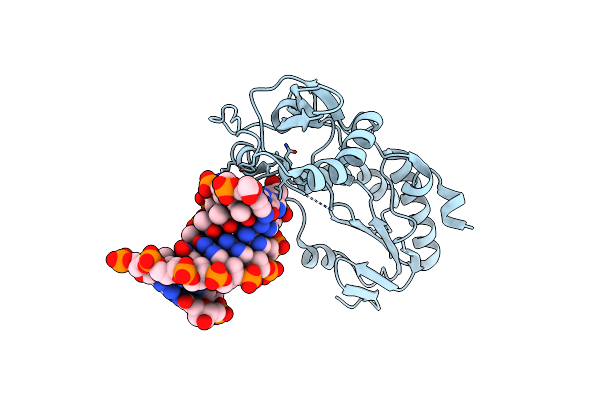 |
Crystal Structure Of B-Form Alien Dna 5'-Cttsspbzpszbbaag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-10 Classification: DNA BINDING PROTEIN/DNA |
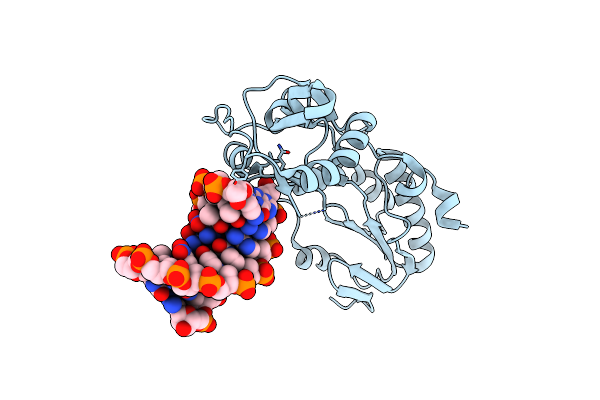 |
Crystal Structure Of B-Form Alien Dna 5'-Cttzzpbsbszppaag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-05-10 Classification: DNA BINDING PROTEIN/DNA |
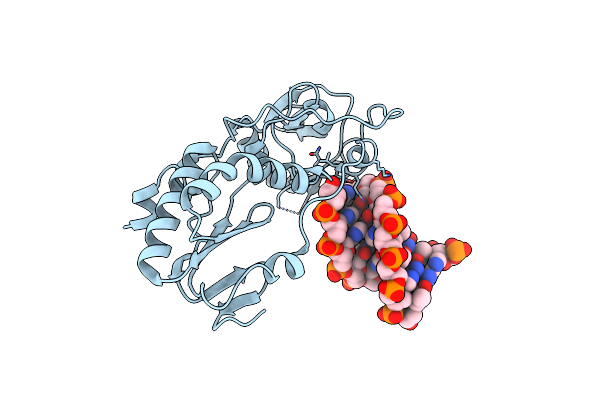 |
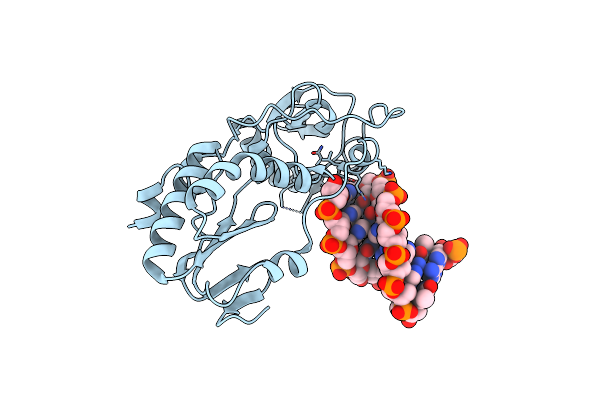
Organism: Moloney murine leukemia virus (isolate shinnick), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-27 Classification: HYDROLASE/DNA |
 |
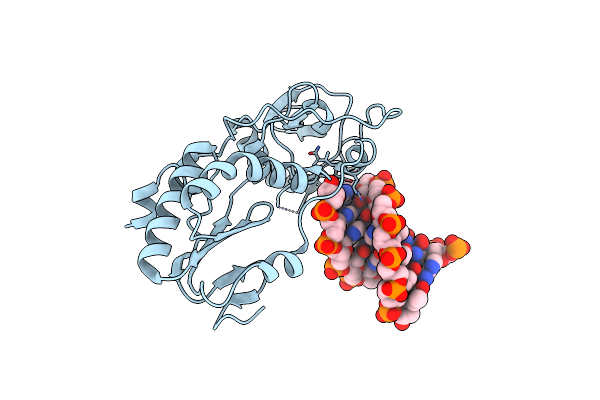
Organism: Moloney murine leukemia virus (isolate shinnick), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-02-27 Classification: HYDROLASE/DNA |
 |
Organism: Moloney murine leukemia virus (isolate shinnick), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-27 Classification: HYDROLASE/DNA |
 |
Hydrogen Bonding Complementary, Not Size Complementarity Is Key In The Formation Of The Double Helix
Organism: Moloney murine leukemia virus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-19 Classification: DNA BINDING PROTEIN/DNA |
 |
Hydrogen Bonding Complementary, Not Size Complementarity Is Key In The Formation Of The Double Helix
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-09-19 Classification: DNA BINDING PROTEIN/DNA |
 |
Hydrogen Bonding Complementary, Not Size Complementarity Is Key In The Formation Of The Double Helix
Organism: Moloney murine leukemia virus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-19 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Thermus aquaticus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2018-07-18 Classification: DNA binding protein/DNA Ligands: A5J, MG |
 |
Structural Basis For Recognition Of Artificial Dna By An Evolved Klentaq Variant
Organism: Thermus aquaticus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2018-07-18 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Moloney murine leukemia virus (isolate shinnick), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-03-07 Classification: transferase/dna |

