Search Count: 36
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: AGS, MG |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: AGS, MG |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: AGS, MG |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: AGS, MG |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: AGS, MG |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: AGS, MG, ZN |
 |
Primase Of Mutant Bacteriophage T4 Primosome With Single Strand Dna/Rna Primer Hybrid In Primer Exiting State
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION/DNA/RNA Ligands: ZN |
 |
Mutant Bacteriophage T4 Gp41 Helicase Hexamer Bound With Single Strand Dna And Atpgammas In The Stalled Primosome
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: REPLICATION Ligands: AGS, MG |
 |
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: REPLICATION Ligands: AGS, MG, ZN |
 |
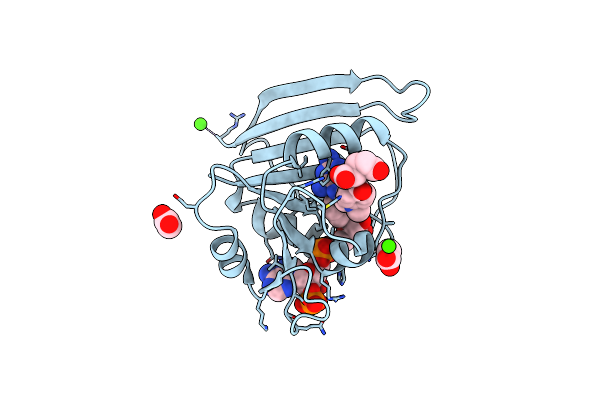
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: MTX, NAP, CA |
 |
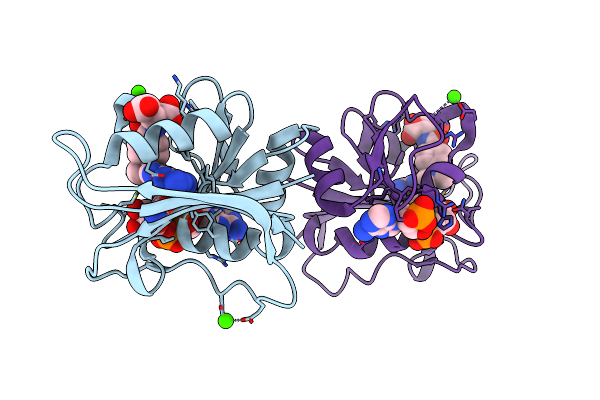
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE Ligands: MTX, NAP, ACT, CA |
 |
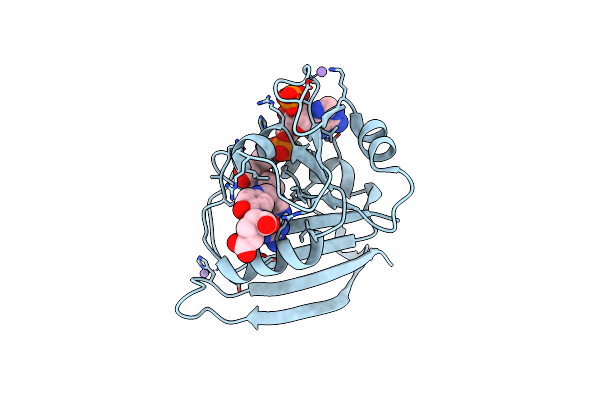
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-06-05 Classification: OXIDOREDUCTASE Ligands: MTX, NDP, CA |
 |
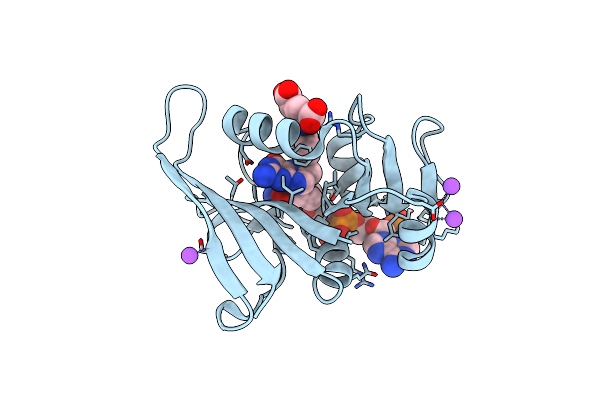
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-04-27 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, NA |
 |
Crystal Structure Of Streptococcus Pneumoniae Dihydrofolate Reductase - Sp9 Mutant
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-02-02 Classification: OXIDOREDUCTASE Ligands: NDP, MTX |
 |
Leucyl-Trna Synthetase From Thermus Thermophilus Complexed With A Sulphamoyl Analogue Of Leucyl-Adenylate In The Synthetic Site And An Adduct Of Amp With 5-Fluoro-1,3-Dihydro-1-Hydroxy-2,1-Benzoxaborole (An2690) In The Editing Site
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-07-03 Classification: LIGASE Ligands: LEU, LMS, ZN, ANZ, SO4 |
 |
Leucyl-Trna Synthetase From Thermus Thermophilus Complexed With A Trna(Leu) Transcript With 5-Fluoro-1,3-Dihydro-1-Hydroxy-2,1- Benzoxaborole (An2690) Forming An Adduct To The Ribose Of Adenosine- 76 In The Enzyme Editing Site.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2007-07-03 Classification: LIGASE Ligands: LEU, ZN, HG, SO4 |
 |
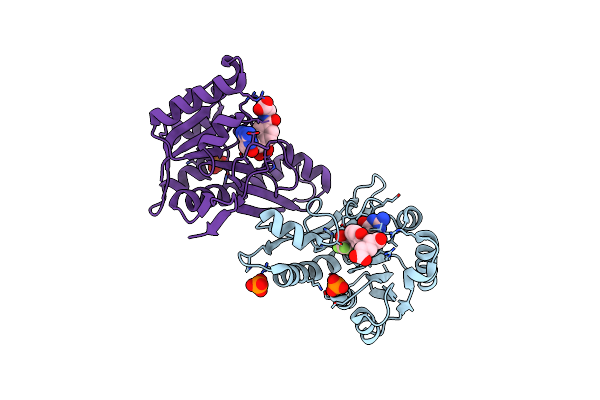
Crystal Structure Of The Homodimeric Bifunctional Transformylase And Cyclohydrolase Enzyme Avian Atic In Complex With A Multisubstrate Adduct Inhibitor Beta-Dadf.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-09-30 Classification: TRANSFERASE, HYDROLASE Ligands: K, PO4, MS1 |
 |
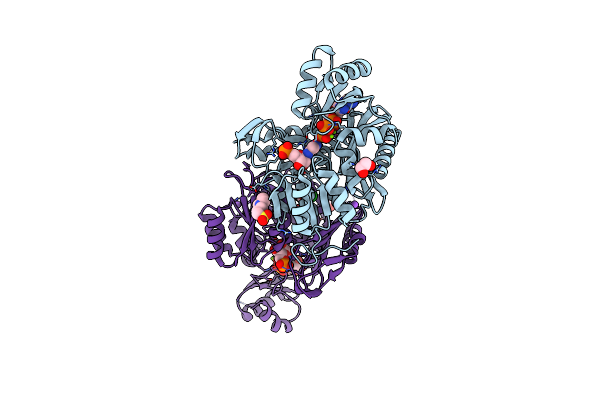
Human Gar Tfase In Complex With Hydrolyzed Form Of 10-Trifluoroacetyl-5,10-Dideaza-Acyclic-5,6,7,8-Tetrahydrofolic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2003-06-10 Classification: TRANSFERASE Ligands: PO4, KEU |
 |
Crystal Structure Of Purt-Encoded Glycinamide Ribonucleotide Transformylase In Complex With Mg-Atp And Gar
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2002-06-28 Classification: TRANSFERASE Ligands: MG, NA, CL, ATP, GAR, MPO, EDO |

