Search Count: 98
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Thermococcus gorgonarius
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-10-02 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-09-11 Classification: CELL CYCLE |
 |
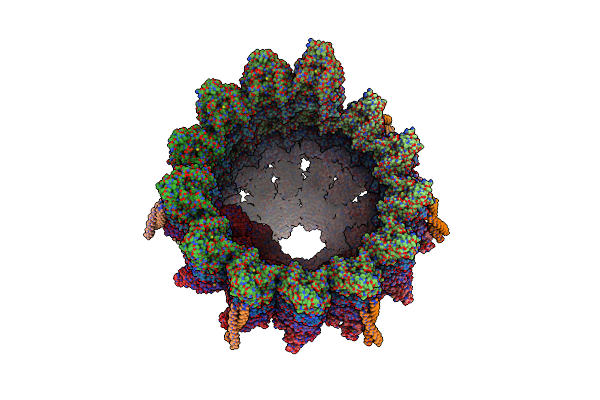
Structure Of The Native Microtubule Lattice Nucleated From The Yeast Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE |
 |
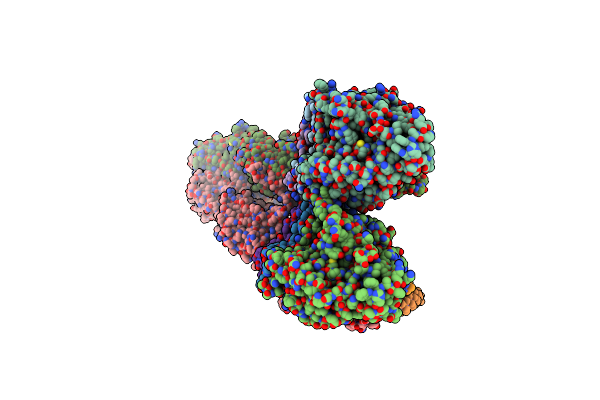
Structure Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP, GDP |
 |
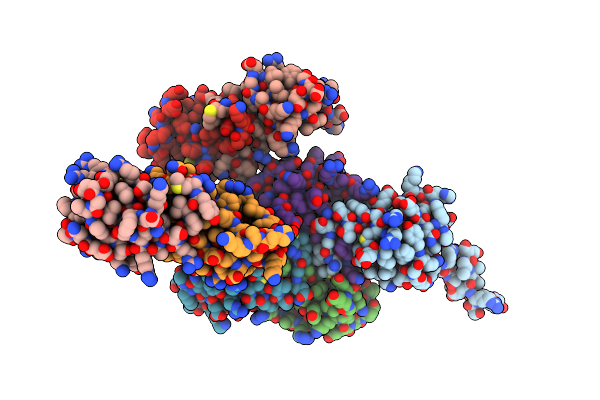
Structure Of The Y-Tubulin Small Complex (Ytusc) As Part Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP |
 |
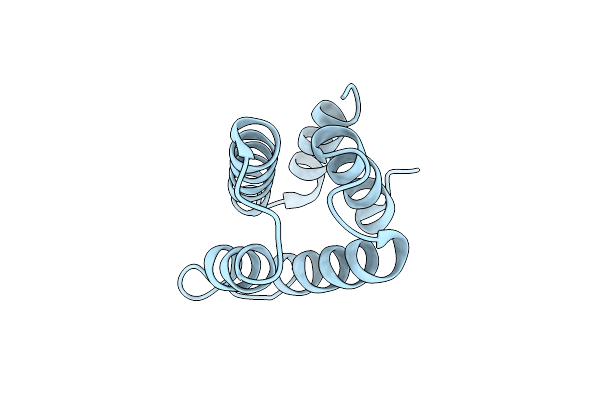
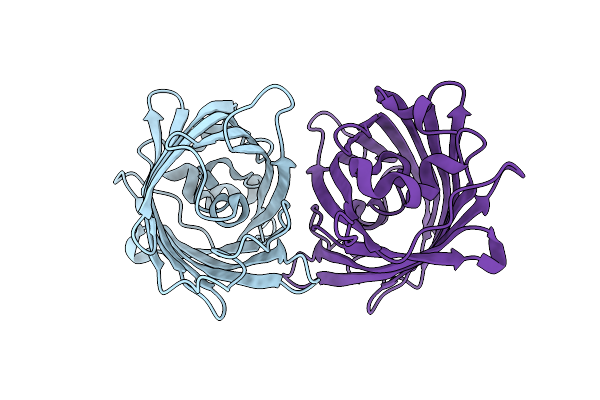
Crystal Structure Of The Hetero-Dimeric Complex From Archaeoglobus Fulgidus Prc1 And Prc2 Domains
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-04-10 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-04-03 Classification: CELL CYCLE |
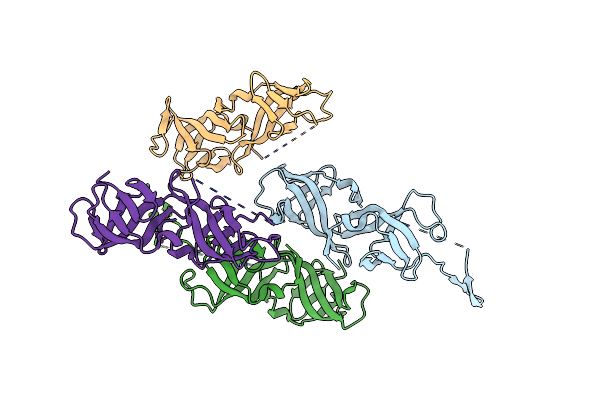 |
Crystal Structure Of Heterodimeric Complex Of Cdpb1 And Cdpb2 From A. Fulgidus
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-04-03 Classification: CELL CYCLE |
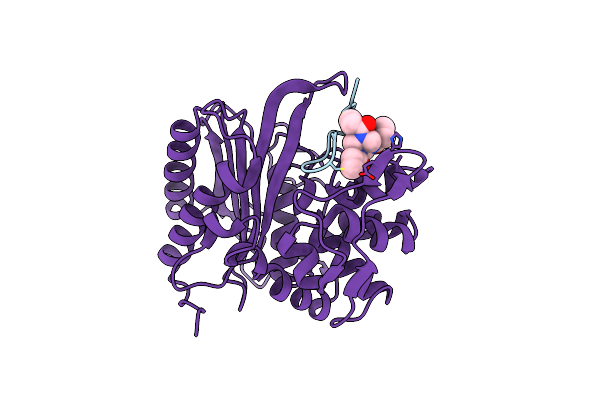 |
The Structure Of E. Coli Penicillin Binding Protein 3 (Pbp3) In Complex With A Bicyclic Peptide Inhibitor
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-04-03 Classification: HYDROLASE Ligands: 29N |
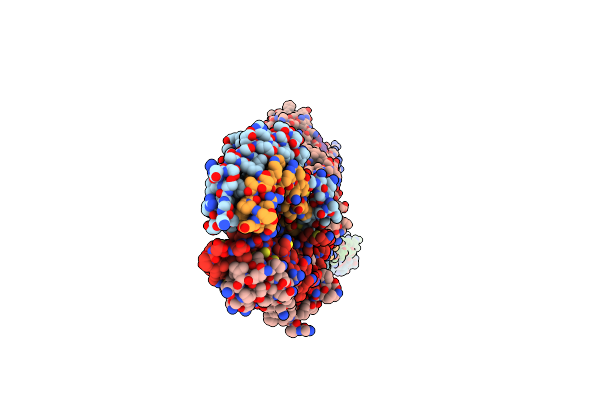 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: CELL CYCLE Ligands: HOH |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-01-10 Classification: TRANSLATION |
 |
Crystal Structure Of Glc7 Phosphatase In Complex With The Regulatory Region Of Ref2
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-29 Classification: GENE REGULATION Ligands: MN, PO4 |
 |
Crystal Structure Of The P110Alpha Catalytic Subunit From Homo Sapiens In Complex With Activator 1938
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2023-05-24 Classification: TRANSFERASE Ligands: QIH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2023-02-15 Classification: TRANSFERASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-01 Classification: TRANSLATION Ligands: MN, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: PO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: 1LT, NA |
 |
Organism: Odinarchaeota archaeon (strain lcb_4)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-06-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: CELL CYCLE |

