Search Count: 7
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-11-28 Classification: CELL ADHESION Ligands: GOL |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2018-11-28 Classification: CELL ADHESION Ligands: NAG, ACT, PEG |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-11-28 Classification: CELL ADHESION Ligands: NAG, PEG, EDO, ACT |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2018-11-28 Classification: CELL ADHESION Ligands: FLC, PEG, BU3, NAG |
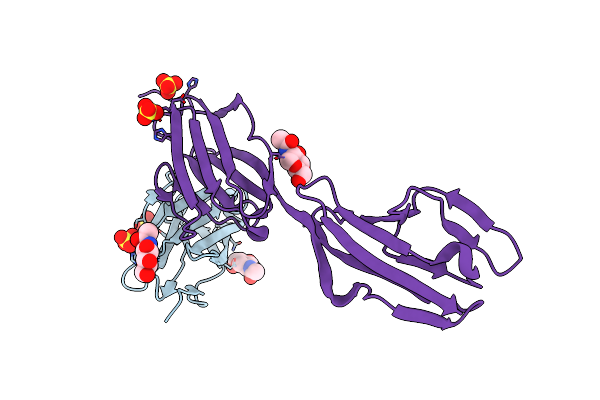 |
Crystal Structure Of The Complex Of Dpr6 Domain 1 Bound To Dip-Alpha Domain 1+2
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-01-06 Classification: CELL ADHESION Ligands: NAG, SO4 |
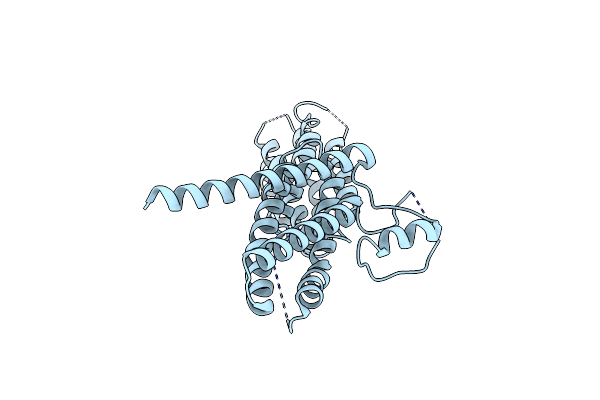 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-09-13 Classification: PROTEIN TRANSPORT |
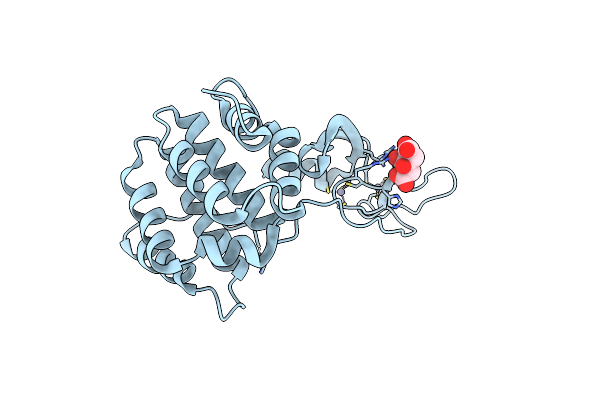 |
Crystal Structure Of The Vhs And Fyve Tandem Domains Of Hrs, A Protein Involved In Membrane Trafficking And Signal Transduction
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-03-06 Classification: TRANSFERASE Ligands: ZN, CIT |

