Search Count: 16
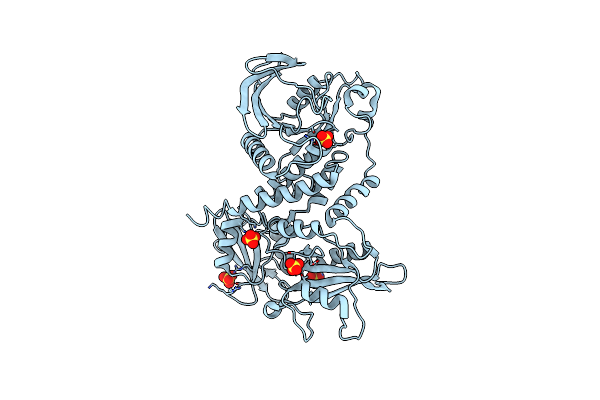 |
Crystal Structure Of The Full-Length Human Protein Tyrosine Phosphatase Shp-1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2011-04-20 Classification: HYDROLASE, SIGNALING PROTEIN Ligands: SO4 |
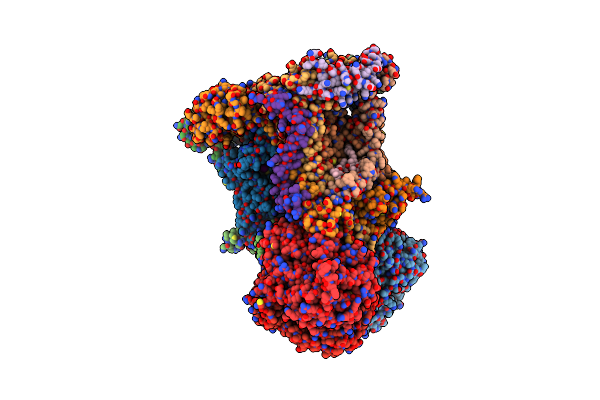 |
Chicken Cytochrome Bc1 Complex With Zn++ And An Iodinated Derivative Of Kresoxim-Methyl Bound
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2009-04-28 Classification: OXIDOREDUCTASE Ligands: PEE, UNL, HEM, IKR, UQ, CDL, ZN, GOL, HEC, BOG, FES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-24 Classification: MEMBRANE PROTEIN Ligands: TRS |
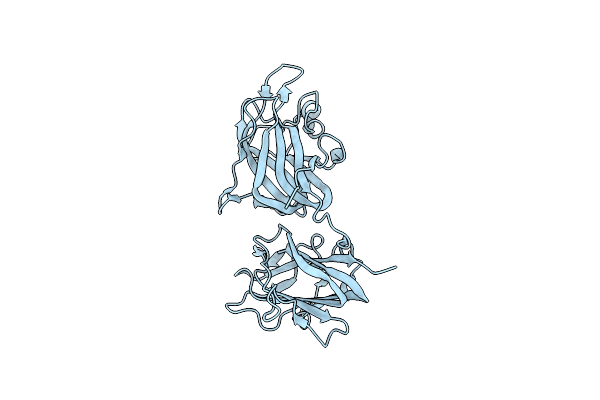 |
Structural Basis For Ligand Binding And Heparin Mediated Activation Of Neuropilin
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-04-03 Classification: SIGNALING PROTEIN, MEMBRANE PROTEIN |
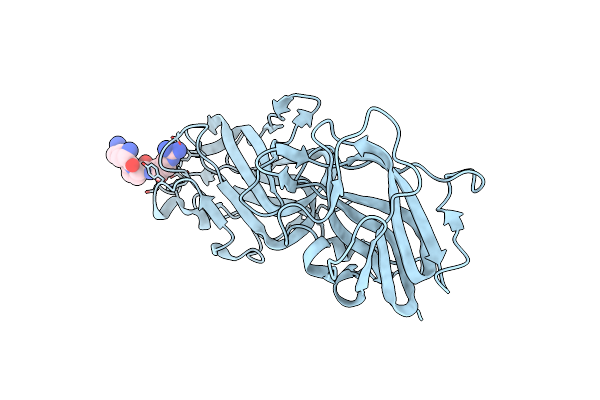 |
Structural Basis For Ligand Binding And Heparin Mediated Activation Of Neuropilin
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2007-04-03 Classification: SIGNALING PROTEIN, MEMBRANE PROTEIN |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dehydrogenase And Azurin From Alcaligenes Faecalis (Form 3)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dephydrogenase And Azurin From Alcaligenes Faecalis (Form 1)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dephydrogenase And Azurin From Alcaligenes Faecalis (Form 2)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-10-17 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of The C2 Domain Of Class Ii Phosphatidylinositide 3-Kinase C2
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-12-13 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-01-20 Classification: CHAPERONE Ligands: ZN |
 |
Structure At 1.9 A Resolution Of A Quinohemoprotein Alcohol Dehydrogenase From Pseudomonas Putida Hk5
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-07-10 Classification: OXIDOREDUCTASE Ligands: CA, PQQ, HEC, EPE, ACN, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2000-05-24 Classification: LIPID BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2000-04-10 Classification: LIPID BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1998-11-11 Classification: LIPID BINDING PROTEIN |
 |
Improvement Of The 2.5 Angstroms Resolution Model Of Cytochrome B562 By Redetermining The Primary Structure And Using Molecular Graphics
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 1991-01-15 Classification: ELECTRON TRANSPORT Ligands: SO4, HEM |

