Search Count: 527
 |
Organism: Saccharomyces cerevisiae, Inovirus m13
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Cryo-Em Structure Of Yeast Mgm101 Bound To Duplex Dna Annealing Intermediate
Organism: Saccharomyces cerevisiae, Inovirus m13
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae, Inovirus m13
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC Ligands: MG |
 |
Crystal Structure Of Salmonella Frab Deglycase, Crystal Form 4 With Deletion Of C-Terminal Residues 313-325.
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
Crystal Structure Of Salmonella Frab Deglycase, E214A Mutant, Crystal Form 5
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
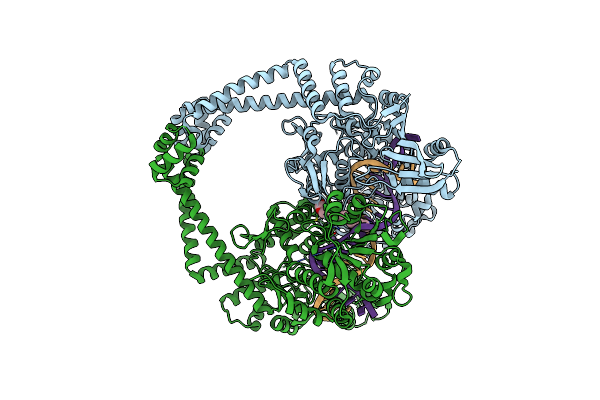
Dna Gyrase Complexed With Uncleaved Dna And Compound 148 To 1.96 A Resolution
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-04-09 Classification: DNA BINDING PROTEIN Ligands: MN, A1BU9 |
 |
Dna Gyrase Complexed With Uncleaved Dna And Compound 185 To 2.25 A Resolution
Organism: Staphylococcus aureus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-04-09 Classification: DNA BINDING PROTEIN Ligands: MN, A1BVQ |
 |
Crystal Structure Of Mycobacterium Tuberculosis Cytochrome P450 Cyp125 In Complex With An Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, A1H47 |
 |
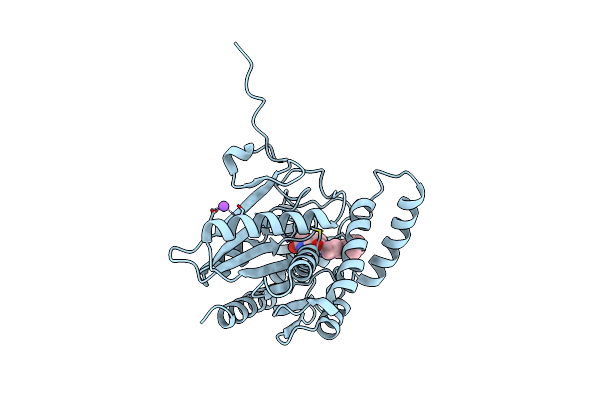
X-Ray Crystal Structure Of Cyp142 From Mycobacterium Tuberculosis In Complex With A Fragment Bound In Two Poses
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: 3QO, HEM |
 |
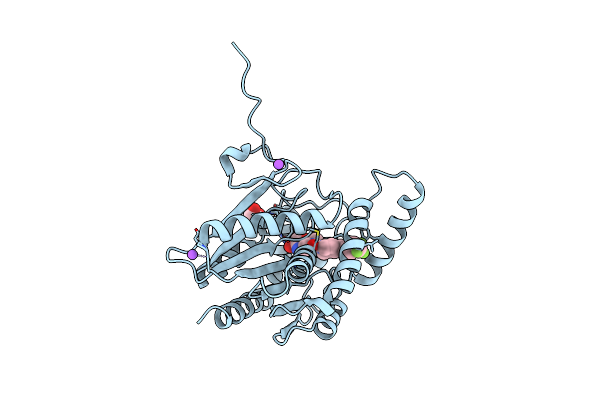
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: A1IA0, NA |
 |
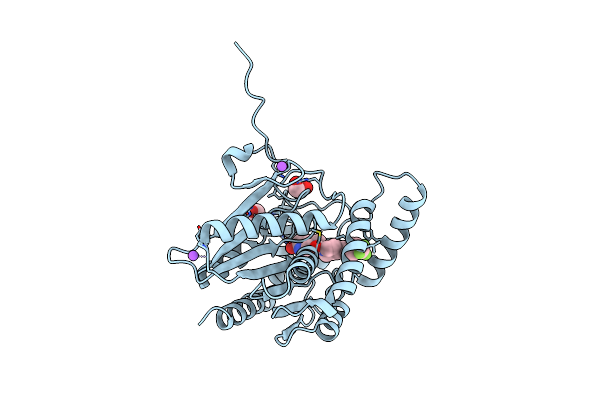
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7N
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-01-22 Classification: HYDROLASE |
 |
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7M
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: A1IA2, NA, EDO |
 |
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7I
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: A1IA1, EDO |
 |
Structure Of Mycobacterium Abscessus Phosphopantetheine Adenylyltransferase In Complex With Fragment
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: 0IM |

