Search Count: 207
 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: SO4, NA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK3, GOL |
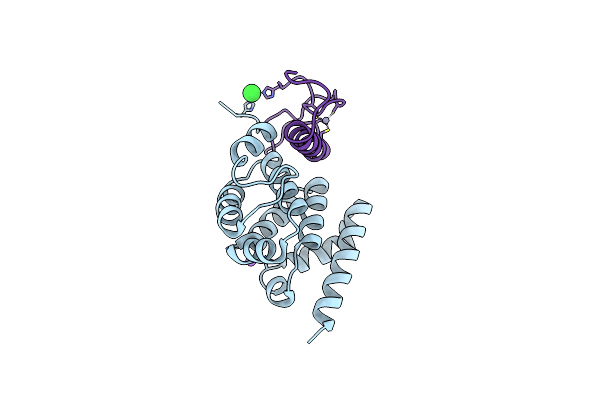 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-05-21 Classification: SIGNALING PROTEIN Ligands: CL, ZN, NA |
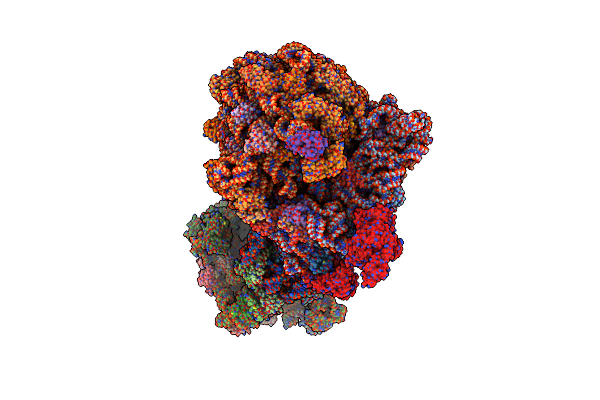 |
Organism: Escherichia coli, Vibrio alginolyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: RIBOSOME |
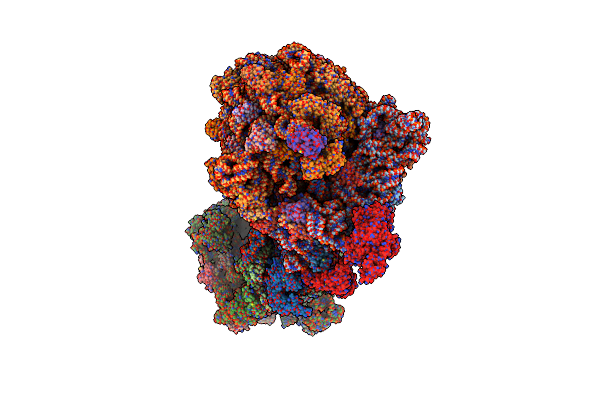 |
Organism: Escherichia coli, Vibrio alginolyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: RIBOSOME |
 |
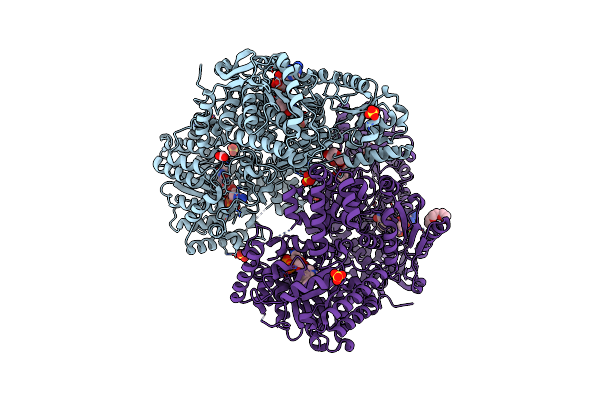
Minimal Puta Proline Dehydrogenase Domain (Design #2) Covalently Inactivated By (S)-But-3-Yn-2-Ylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: A1BC6, FMT |
 |
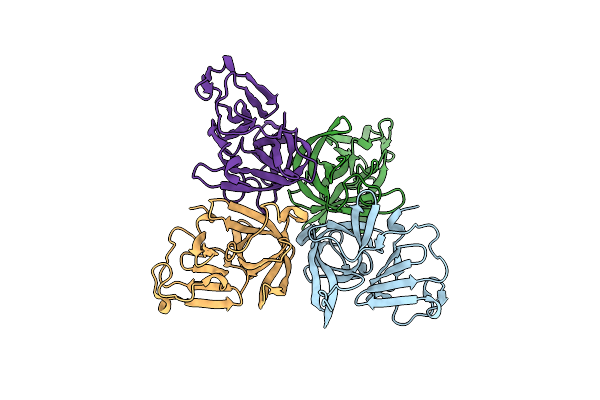
Proline Utilization A (Puta) From Sinorhizobium Meliloti Inactivated By N-Propargylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: P5F, NAD, SO4, MG, FMT, PEG, PG4, PGE |
 |
Structure Of Norovirus (Hu/Gii.4/Sydney/Nsw0514/2012/Au) Protease In The Ligand-Free State
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2024-01-31 Classification: VIRAL PROTEIN |
 |
Organism: Aspergillus fumigatus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-24 Classification: REPLICATION |
 |
Organism: Aspergillus fumigatus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-24 Classification: REPLICATION |
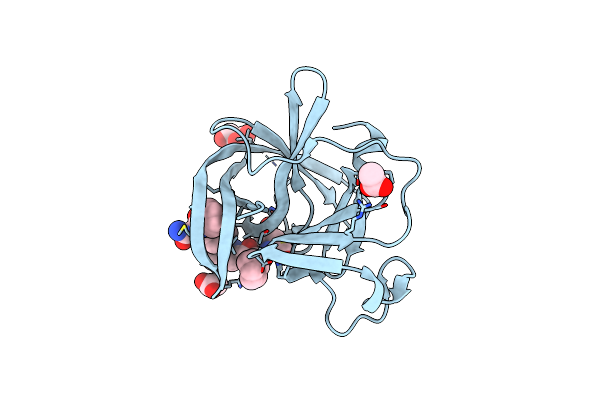 |
Structure Of Norovirus (Hu/Gii.4/Sydney/Nsw0514/2012/Au) Protease Bound To Inhibitor Nv-004
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN/INHIBITOR Ligands: GOL, ACT, FHR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-05-10 Classification: TRANSCRIPTION Ligands: A8R |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-05-10 Classification: TRANSCRIPTION Ligands: A8R |
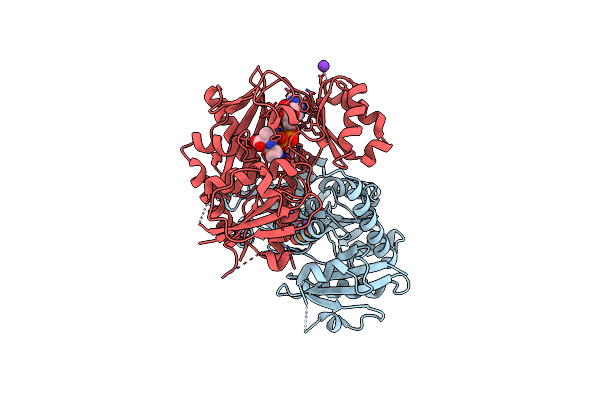 |
Staphylococcus Aureus D-Alanine-D-Alanine Ligase In Complex With Atp, D-Ala-D-Ala, Mg2+ And K+
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-15 Classification: LIGASE Ligands: K, MG, ADP, ATP |
 |
Crystal Structure Of Staphylococcus Aureus Biotin Protein Ligase In Complex With Inhibitor
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-11-30 Classification: LIGASE/LIGASE INHIBITOR Ligands: WNE |
 |
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-09-14 Classification: LIGASE Ligands: ADP, MG, NA, CL |
 |
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: LIGASE Ligands: CIT, ADP, MG, NA |

