Search Count: 16
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ROC, GOL |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2011-12-07 Classification: HYDROLASE Ligands: ZN, BEN, MPD, IMD |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2011-10-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: RIT, DMS, 1PE, PGE, GOL |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: BEN, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-07-13 Classification: TRANSFERASE Ligands: IPH |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-05-25 Classification: HYDROLASE Ligands: XE, GOL |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2011-05-25 Classification: HYDROLASE Ligands: ZN, CA, PGO |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2011-05-25 Classification: ISOMERASE Ligands: MN, CL, PGO |
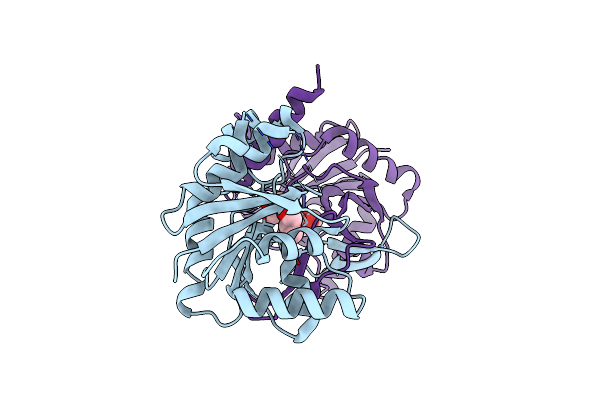 |
Crystal Structure Of 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase (Ispd) In Complex With 1,2-Propanediol
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-05-25 Classification: TRANSFERASE Ligands: PGO, PGR |
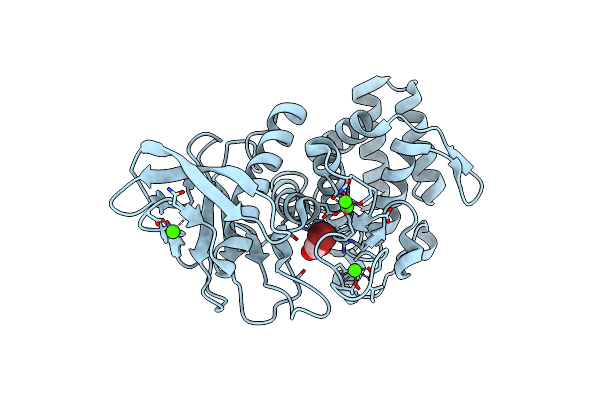 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-05-25 Classification: HYDROLASE Ligands: ZN, CA, BXA |
 |
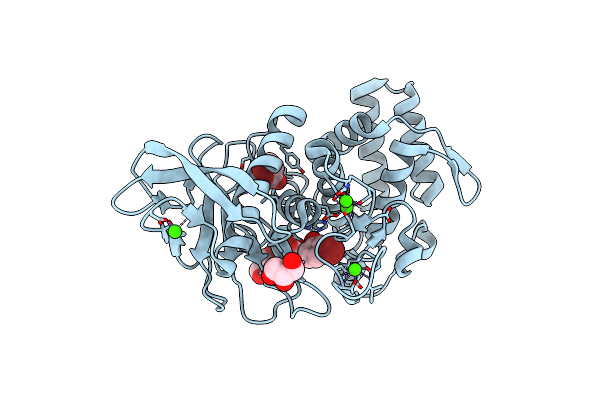
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2011-05-11 Classification: HYDROLASE Ligands: ZN, CA, ANL, IPA, GOL, DMS |
 |
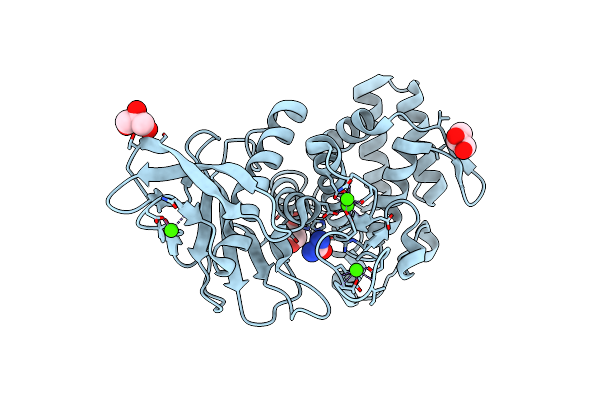
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2011-05-11 Classification: HYDROLASE Ligands: B3R, ZN, CA, GOL, IPA |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-05-11 Classification: HYDROLASE Ligands: ZN, CA, URE, GOL |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2011-05-11 Classification: HYDROLASE Ligands: ZN, CA, NMU, GOL |
 |
Organism: Torpedo californica
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2011-03-09 Classification: HYDROLASE Ligands: XE, NAG, PG4, PEG |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2011-02-23 Classification: HYDROLASE Ligands: XE, CA, ZN, VAL, LYS, GOL, DMS |

