Search Count: 137
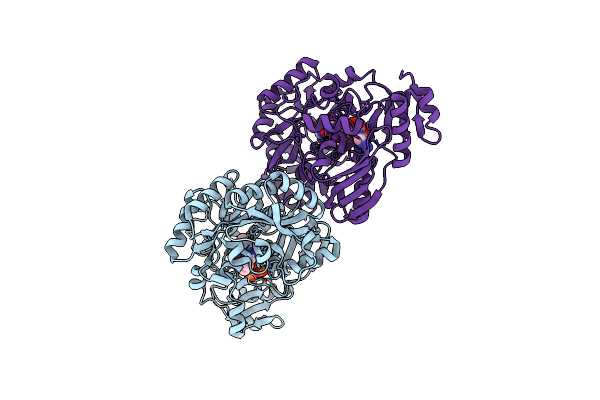 |
Crystal Structure Of The Flavin Oxygenase With Cofactor And Substrate Bound Involved In Folate Catabolism
Organism: Hoeflea phototrophica (strain dsm 17068 / ncimb 14078 / dfl-43)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-12-09 Classification: OXIDOREDUCTASE Ligands: LUZ, FAD |
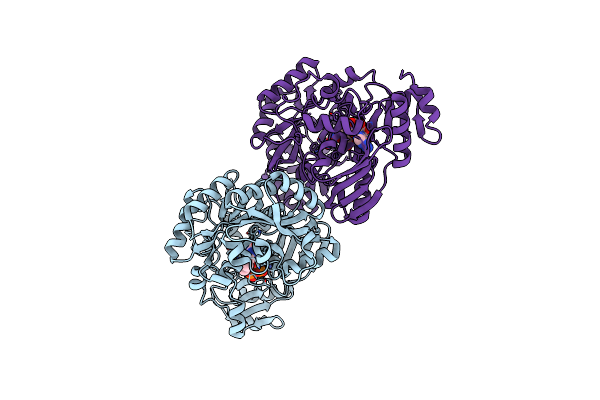 |
Crystal Structure Of The Flavin Oxygenase With Cofactor Bound Involved In Folate Catabolism
Organism: Hoeflea phototrophica (strain dsm 17068 / ncimb 14078 / dfl-43)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: FAD |
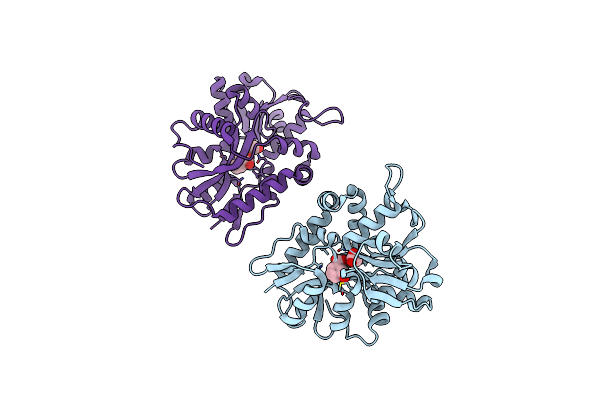 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2019-04-03 Classification: LYASE Ligands: ACT, 3HB |
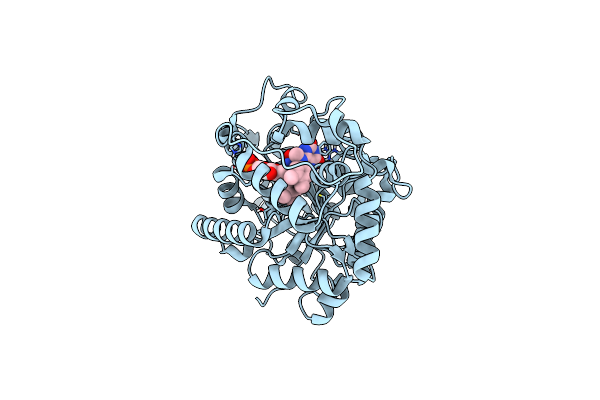 |
Crystal Structure Of Flavin Monooxygenase Cmoj (Earlier Ytnj) Bound With Fmn
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-12 Classification: FLAVOPROTEIN Ligands: FMN, LFN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-08-29 Classification: FLAVOPROTEIN, OXIDOREDUCTASE |
 |
Organism: Herbiconiux
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-06-13 Classification: FLAVOPROTEIN Ligands: SO4 |
 |
Organism: Herbiconiux
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-06-13 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of Riboflavin Lyase (Rcae) With Modified Fmn And Substrate Riboflavin
Organism: Herbiconiux
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-06-13 Classification: FLAVOPROTEIN Ligands: 9WY, RBF |
 |
Crystal Structure Of Burkholderia Thailandensis 1,6-Didemethyltoxoflavin-N1-Methyltransferase With Bound 1,6-Didemethyltoxoflavin And S-Adenosylhomocysteine
Organism: Burkholderia thailandensis (strain atcc 700388 / dsm 13276 / cip 106301 / e264)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2017-12-13 Classification: TRANSFERASE Ligands: SAH, AZ8, SO4, EPE |
 |
Crystal Structure Of Burkholderia Thailandensis 1,6-Didemethyltoxoflavin-N1-Methyltransferase With Bound S-Adenosylhomocysteine
Organism: Burkholderia thailandensis (strain atcc 700388 / dsm 13276 / cip 106301 / e264)
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2017-12-13 Classification: TRANSFERASE Ligands: SAH, SO4, EPE |
 |
Crystal Structure Of A Flavoenzyme Ruta In The Pyrimidine Catabolic Pathway
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-19 Classification: OXIDOREDUCTASE Ligands: URA, SO4, GOL, FMN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2017-01-18 Classification: LYASE Ligands: PLP, GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2017-01-18 Classification: LYASE Ligands: 5RP, PO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-01-18 Classification: LYASE Ligands: KPR, PO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2017-01-18 Classification: LYASE Ligands: PO4, SO4, KIK |
 |
Crystal Structure Of Arabidopsis Thaliana Pdx1-I320 Complex From Multiple Crystals
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2017-01-18 Classification: LYASE Ligands: PO4, SO4, KIK |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-18 Classification: LYASE Ligands: K8P, RP5, HG3, GOL |
 |
Crystal Structure Of Burkholderia Glumae Toxa Y7F Mutant With Bound S-Adenosylhomocysteine (Sah)
Organism: Burkholderia glumae
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2016-05-11 Classification: TRANSFERASE Ligands: SAH, DMS, PG4, TRS |
 |
Organism: Burkholderia glumae
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2016-05-11 Classification: TRANSFERASE Ligands: EDO, GOL |
 |
Crystal Structure Of Burkholderia Glumae Toxa Y7F Mutant With Bound S-Adenosylhomocysteine (Sah) And Toxoflavin
Organism: Burkholderia glumae
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-05-04 Classification: TRANSFERASE Ligands: SAH, TOF, TRS |

