Search Count: 14
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Analysis Of The Interaction Between Charged Side Chains And The Alpha-Helix Dipole Using Designed Thermostable Mutants Of Phage T4 Lysozyme
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: BME |
 |
Analysis Of The Interaction Between Charged Side Chains And The Alpha-Helix Dipole Using Designed Thermostable Mutants Of Phage T4 Lysozyme
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) |
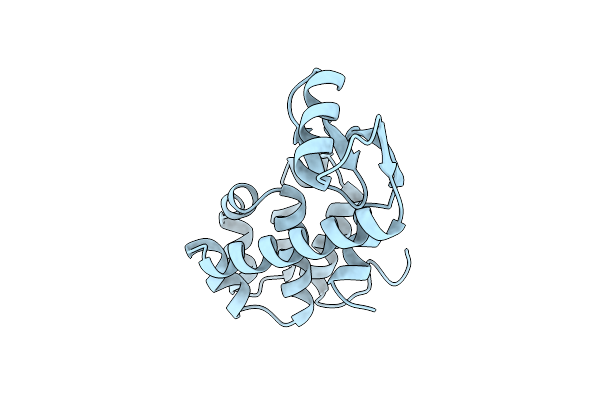 |
Hydrophobic Stabilization In T4 Lysozyme Determined Directly By Multiple Substitutions Of Ile 3
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
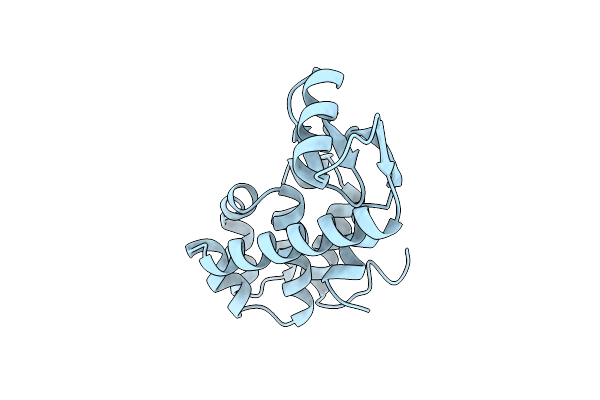 |
Hydrophobic Stabilization In T4 Lysozyme Determined Directly By Multiple Substitutions Of Ile 3
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
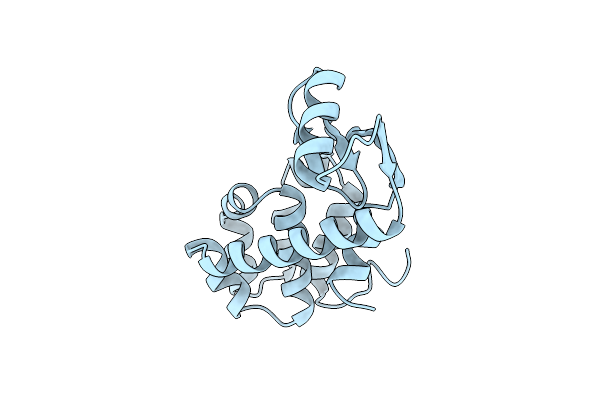 |
Enhanced Protein Thermostability From Designed Mutations That Interact With Alpha-Helix Dipoles
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
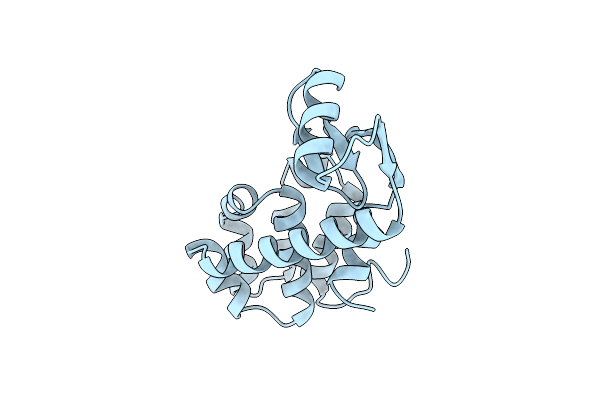 |
Enhanced Protein Thermostability From Designed Mutations That Interact With Alpha-Helix Dipoles
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Enhanced Protein Thermostability From Site-Directed Mutations That Decrease The Entropy Of Unfolding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Enhanced Protein Thermostability From Site-Directed Mutations That Decrease The Entropy Of Unfolding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |

