Search Count: 19
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: RNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: RNA BINDING PROTEIN Ligands: ADP, MG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: RNA BINDING PROTEIN Ligands: ADP, ALF |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: RNA BINDING PROTEIN Ligands: ADP, MG, ALF |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: RNA BINDING PROTEIN Ligands: ADP, ALF, MG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: RNA BINDING PROTEIN Ligands: ADP, ALF |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: RNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl-6.2 (Dna Conformation 1)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl+5.8 (Composite Map)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
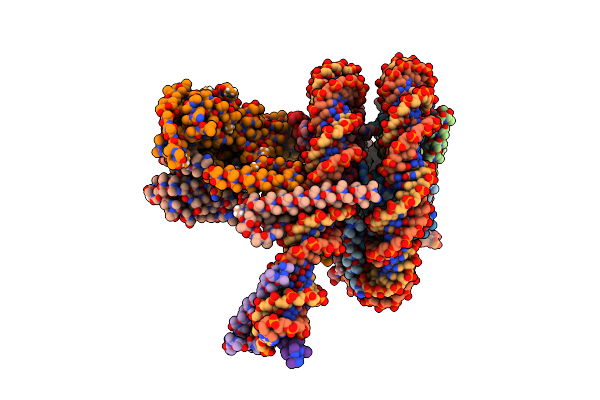 |
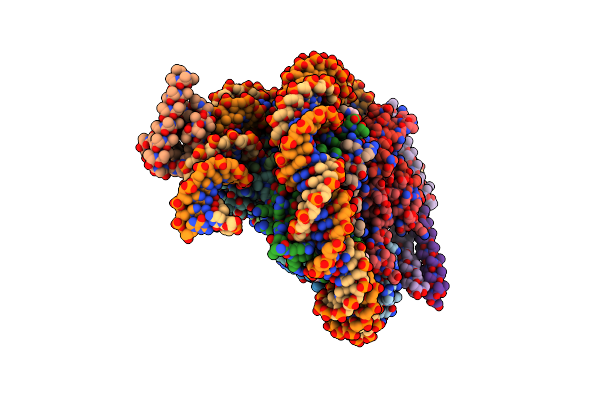
Cryo-Em Structure Of Clock-Bmal1 Bound To The Native Por Enhancer Nucleosome (Map 2, Additional 3D Classification And Flexible Refinement)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
 |
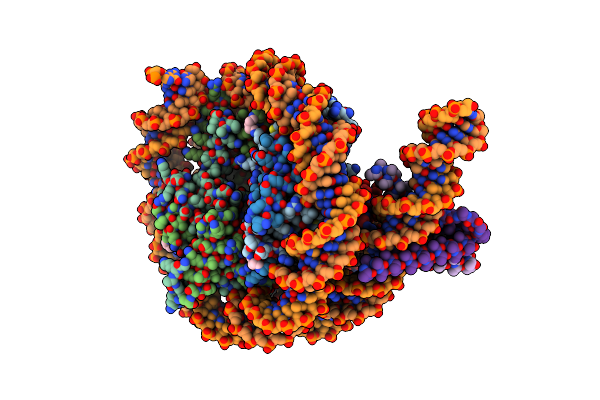
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Structure, Dynamics And Rox2-Lncrna Binding Of Tandem Double-Stranded Rna Binding Domains Dsrbd1/2 Of Drosophila Helicase Mle
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2019-02-20 Classification: RNA BINDING PROTEIN |
 |
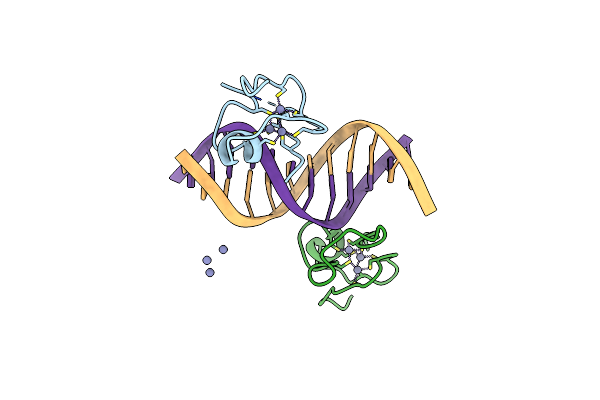
Organism: Drosophila melanogaster, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2015-11-18 Classification: HYDROLASE/RNA Ligands: ADP, MG, ALF, GOL |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-01-21 Classification: DNA binding protein/DNA Ligands: ZN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-01-21 Classification: DNA binding protein/DNA Ligands: ZN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-11-09 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-11-09 Classification: DNA BINDING PROTEIN Ligands: CD |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-09-05 Classification: NUCLEAR PROTEIN Ligands: GLC, G4D, GOL |

