Search Count: 34
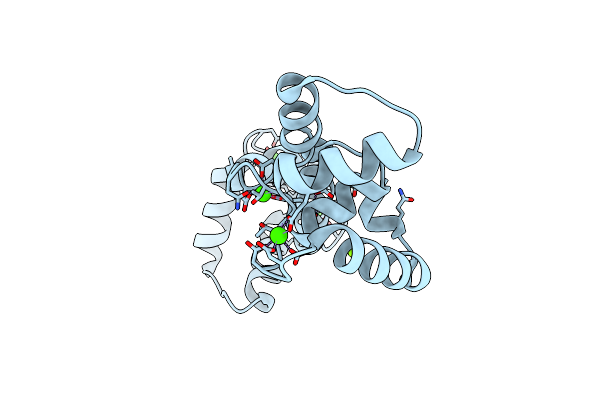 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-10-26 Classification: METAL BINDING PROTEIN Ligands: CA, MG, ZN |
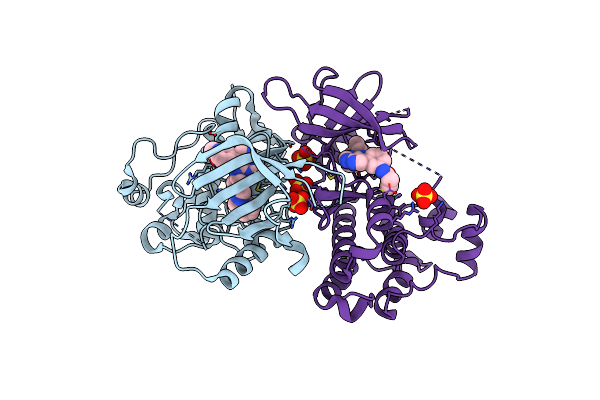 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 14D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7K, SO4 |
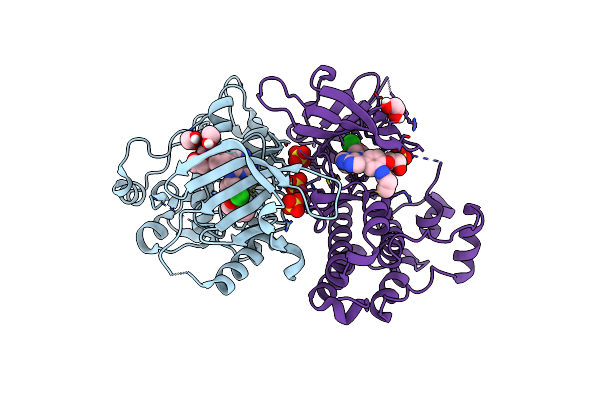 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 18B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7Q, EDO, SO4, CL |
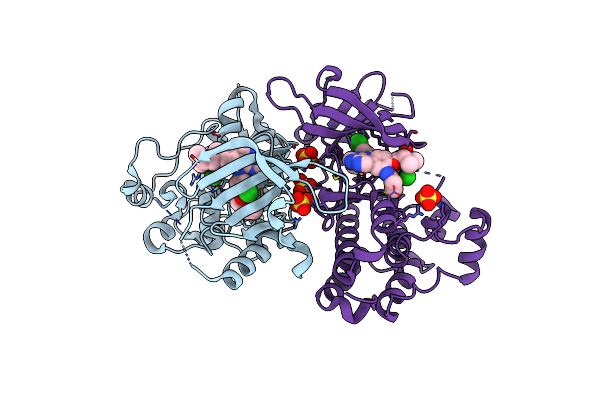 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 18D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7W, SO4, MG, CL |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 18C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7Z, SO4 |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 19
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7B, SO4, EDO |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 21A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N78, EDO, SO4 |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 21B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N82, EDO, SO4, NA |
 |
Crystal Structure Of Glycogen Synthase Kinase-3 Beta (Gsk3B) In Complex With Pik-75
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-06-20 Classification: TRANSFERASE Ligands: CL, F4N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: 6HL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: 6HJ |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-09-09 Classification: TRANSFERASE Ligands: GOL, 56G |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-09-09 Classification: TRANSFERASE Ligands: G97 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: 563 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-05-27 Classification: Transferase/Transferase inhibitor Ligands: 3XM, MES |
 |
Crystal Structure Of Rat Cyclophilin D In Complex With A Potent Nonimmunosuppressive Inhibitor
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2014-11-12 Classification: ISOMERASE/ISOMERASE Inhibitor Ligands: P6G, SO4 |
 |
Crystal Structure Of Human Sirt3 In Complex With Fluor-De-Lys Peptide And Piceatannol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-12-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, PIT, IPA |
 |
Crystal Structure Of Human Sirt5 In Complex With Fluor-De-Lys Peptide And Resveratrol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-12-05 Classification: HYDROLASE/HYDROLASE ACTIVATOR Ligands: ZN, STL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-10-24 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-10-24 Classification: HYDROLASE/HYDROLASE INHIBITOR |

