Search Count: 114
 |
Cryo-Em Structure Of The Spike Glycoprotein From Bat Sars-Like Coronavirus (Bat Sl-Cov) Wiv1 In Locked State
Organism: Bat sars-like coronavirus wiv1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRUS LIKE PARTICLE |
 |
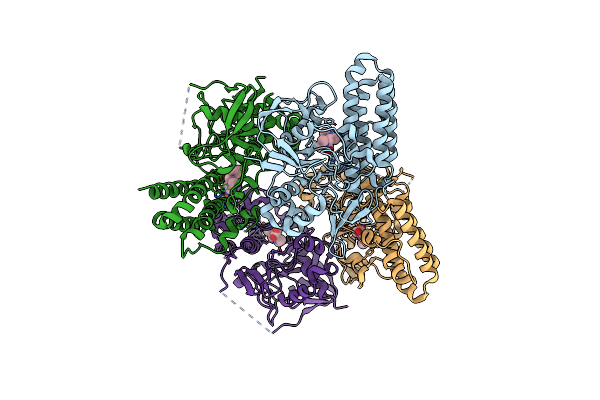
Cryo-Em Structure Of Single-Layered Nucleoprotein-Rna Helical Assembly From Marburg Virus, Trimeric Repeat Unit
Organism: Marburg virus - musoke, kenya, 1980
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
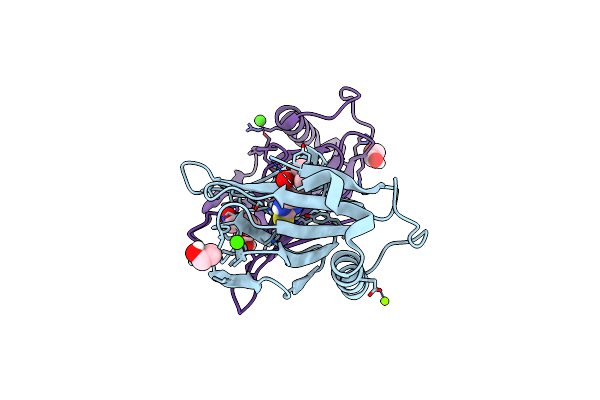
Sulfoxide Synthase From Chloracidobacterium Thermophilum, Egtb Type Ii, In Complex With Iron And Trimethylhistidine
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-11 Classification: OXIDOREDUCTASE Ligands: UCQ, FE |
 |
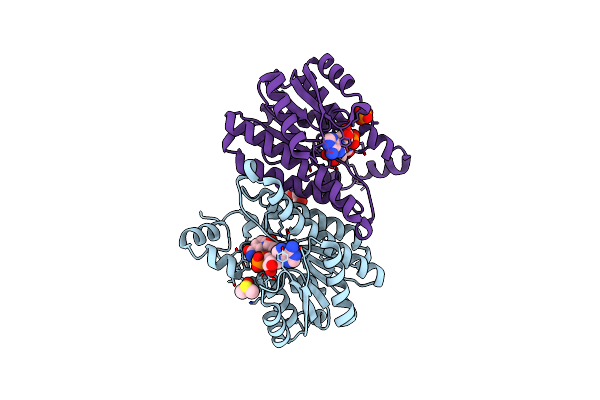
Ergothioneine Dioxygenase From Thermocatellispora Tengchongensis In Complex With Nickel And Substrate (Anaerobic)
Organism: Thermocatellispora tengchongensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-11 Classification: OXIDOREDUCTASE Ligands: EDO, LW8, MG, NI, CA, PG4, ACT |
 |
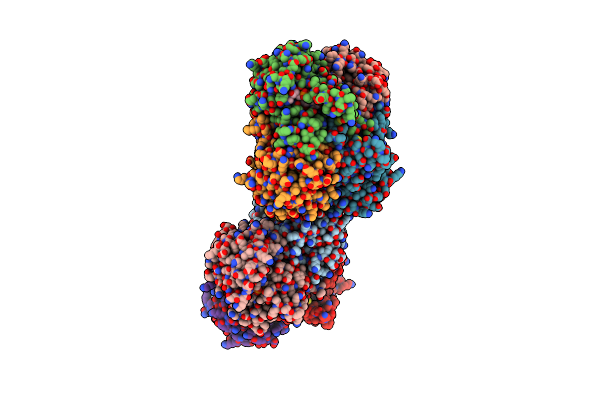
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: NAD, DMS, PEG, PO4 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: NAD, CL |
 |
Cryoem Structure Of A C7-Symmetrical Groel7-Groes7 Cage In Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Disordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel7-Groes7 Cage With Encapsulated Ordered Substrate Metk In The Presence Of Adp-Befx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Structure Average Of Groel14 Complexes Found In The Cytosol Of Escherichia Coli Overexpressing Groel Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
In Situ Structure Average Of Groel14-Groes14 Complexes In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Cryoem Structure Of A Groel14-Groes7 Complex In Presence Of Adp-Befx With Wide Groel7 Trans Ring Conformation
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
Cryoem Structure Of A Groel14-Groes7 Complex In Presence Of Adp-Befx With Narrow Groel7 Trans Ring Conformation
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ADP, MG, BEF, K |
 |
In Situ Structure Average Of Groel14-Groes7 Complexes With Wide Groel7 Trans Ring Conformation In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
In Situ Structure Average Of Groel14-Groes7 Complexes With Narrow Groel7 Trans Ring Conformation In Escherichia Coli Cytosol Obtained By Cryo Electron Tomography
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: CHAPERONE Ligands: ATP, MG, K, ADP |
 |
S-Methylthiourocanate Hydratase, Variant R450A, From Variovorax Sp. Ra8 In Complex With Nad+
Organism: Variovorax sp. ra8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-06 Classification: LYASE Ligands: NAD |
 |
S-Methylthiourocanate Hydratase From Variovorax Sp. Ra8 In Complex With Nad+ And Imidazolone Propionate
Organism: Variovorax sp. ra8
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-06 Classification: OXIDOREDUCTASE Ligands: PG4, O66, NAD, ACY, EDO |
 |
Ergothioneine Dioxygenase From Thermocatellispora Tengchongensis In Complex With Iron
Organism: Thermocatellispora tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-12-27 Classification: OXIDOREDUCTASE Ligands: FE, ACT, MG, PG4 |
 |
Ergothioneine Dioxygenase From Thermocatellispora Tengchongensis In Complex With Manganese
Organism: Thermocatellispora tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-12-27 Classification: OXIDOREDUCTASE Ligands: EDO, PEO, MN, MG, PG4, PEG |

