Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
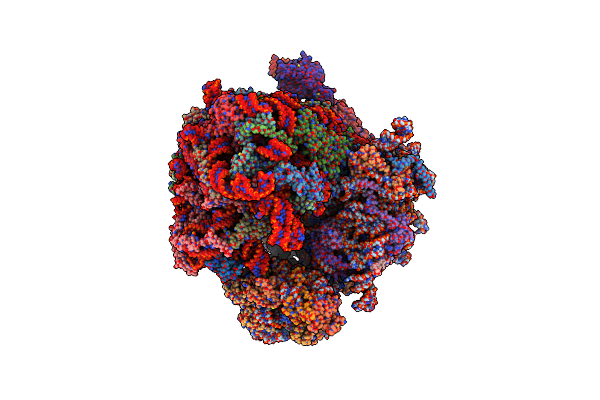 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: RIBOSOME Ligands: ZN |
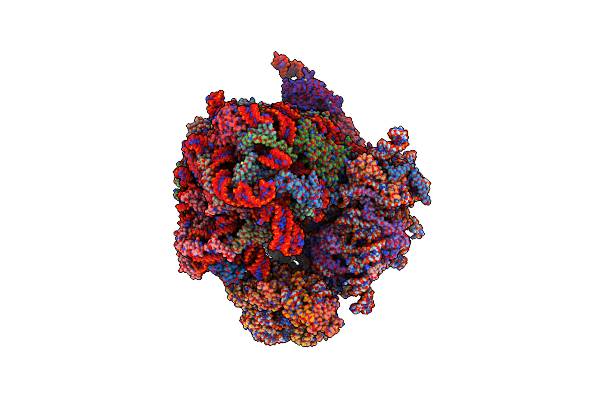 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: RIBOSOME Ligands: ZN |
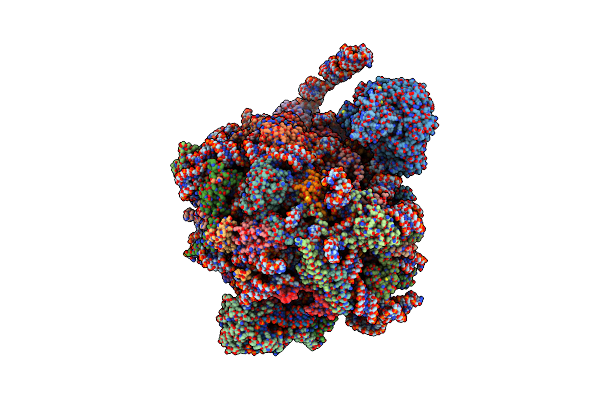 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: RIBOSOME Ligands: MG, ZN |
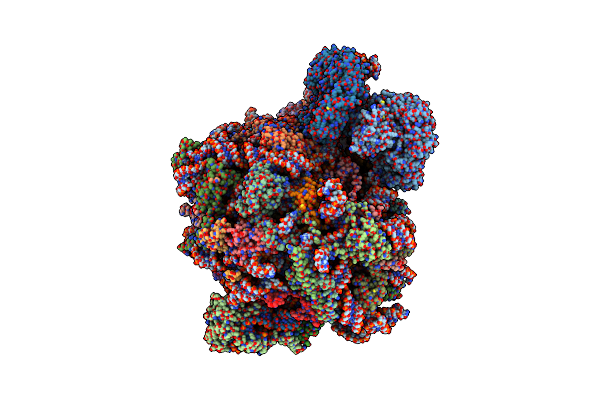 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: RIBOSOME Ligands: MG, ZN |
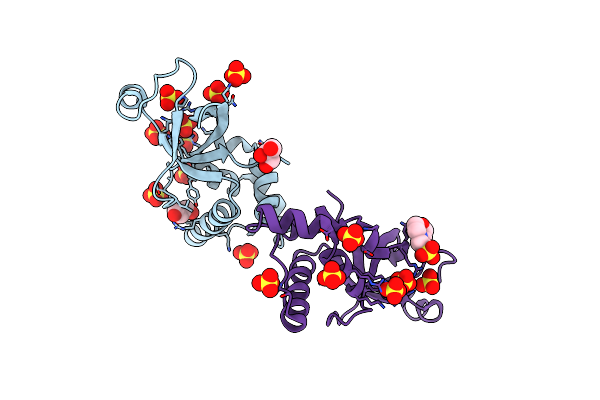 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2020-12-02 Classification: PROTEIN TRANSPORT Ligands: GOL, SO4, MES |
 |
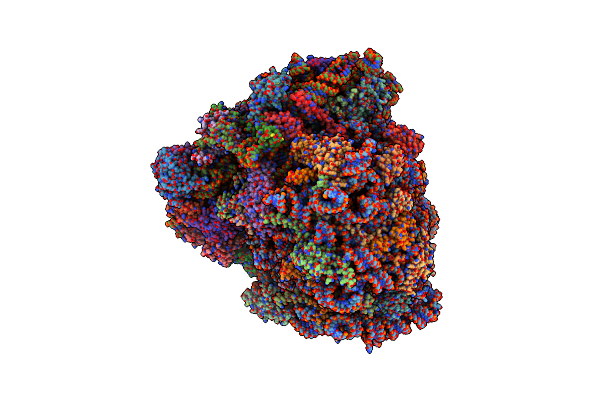
Cryo-Em Structure Of The Signal Sequence-Engaged Post-Translational Sec Translocon
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN |
 |
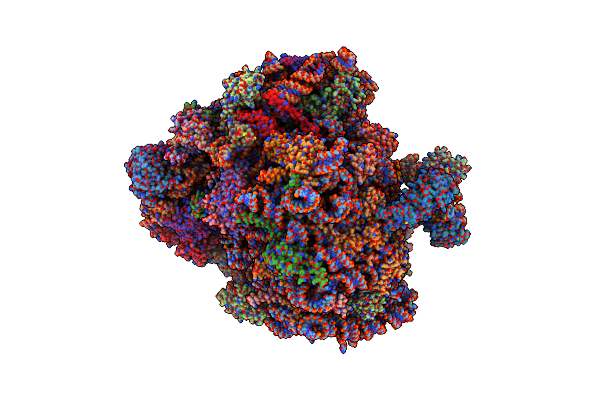
Organism: Homo sapiens, Saccharomyces cerevisiae, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2019-07-10 Classification: RIBOSOME Ligands: MG, ZN |
 |
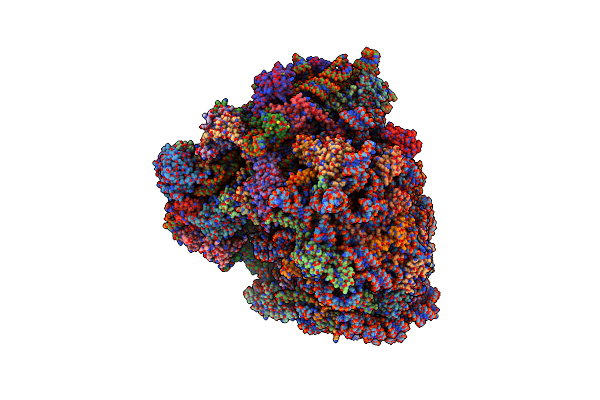
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae, Canis lupus familiaris
Method: ELECTRON MICROSCOPY Release Date: 2019-07-10 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2019-07-10 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae, Canis lupus familiaris
Method: ELECTRON MICROSCOPY Release Date: 2019-07-10 Classification: RIBOSOME Ligands: ZN, MG |
 |
Cryo-Em Structure Of The Mrna Translating And Degrading Yeast 80S Ribosome-Xrn1 Nuclease Complex
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2019-03-13 Classification: TRANSLATION Ligands: ZN, MG |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: TRANSLATION |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2018-12-19 Classification: TRANSLATION Ligands: 3HE |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:7.70 Å Release Date: 2016-01-13 Classification: ISOMERASE |
 |
Localization Of The Small Subunit Ribosomal Proteins Into A 6.1 A Cryo-Em Map Of Saccharomyces Cerevisiae Translating 80S Ribosome
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Organism: Homo sapiens, Escherichia coli, Escherichia coli dh1, Escherichia coli o157:h7, Escherichia coli k-12, Escherichia coli 536
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME/RIBOSOMAL PROTEIN Ligands: PEV, PGV |
 |
Model Of The Small Subunit Rna Based On A 5.5 A Cryo-Em Map Of Triticum Aestivum Translating 80S Ribosome
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
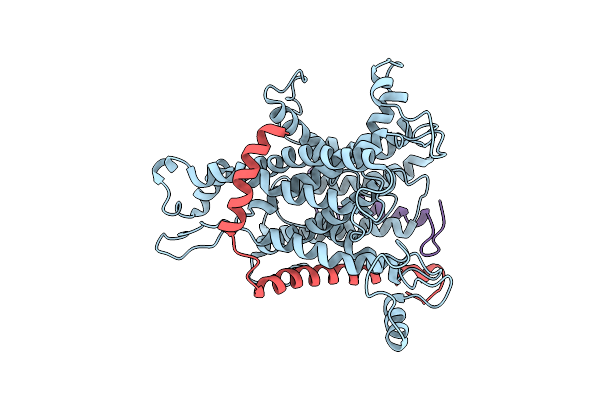 |
Cryo-Em Of The Sec61-Complex Bound To The 80S Ribosome Translating A Secretory Substrate
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:7.40 Å Release Date: 2014-02-05 Classification: PROTEIN TRANSPORT |
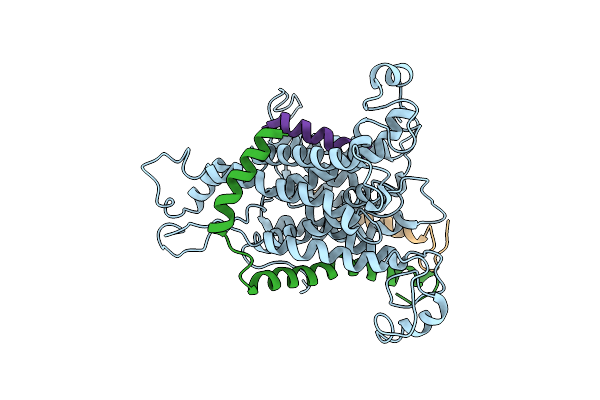 |
Cryo-Em Of The Sec61-Complex Bound To The 80S Ribosome Translating A Membrane-Inserting Substrate
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:7.80 Å Release Date: 2014-02-05 Classification: PROTEIN TRANSPORT |
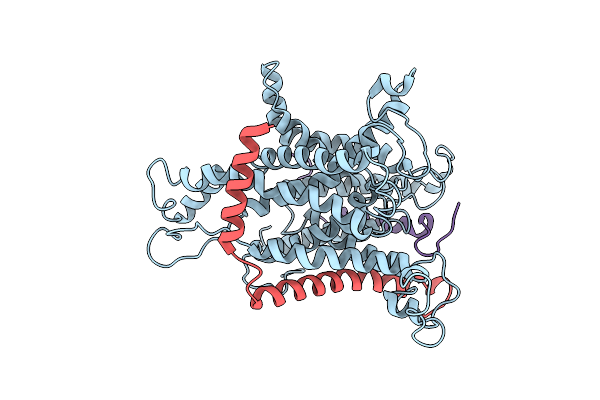 |
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:6.90 Å Release Date: 2014-02-05 Classification: PROTEIN TRANSPORT |