Search Count: 143
 |
Crystal Structure Of Cellulosomal Double-Dockerin Module Of Clo1313_0689 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-03 Classification: PROTEIN BINDING Ligands: CA |
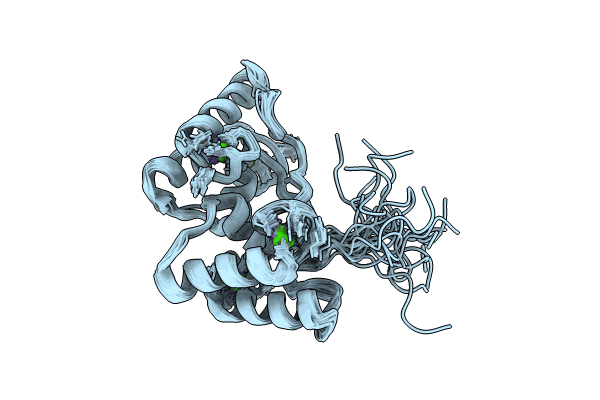 |
Solution Nmr Structure Of Cellulosomal Double-Dockerin Module Of Clo1313_0689 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2024-04-03 Classification: PROTEIN BINDING Ligands: CA |
 |
Clostridium Thermocellum Rna Polymerase Transcription Open Complex With Sigi1 And Its Promoter
Organism: Acetivibrio thermocellus dsm1313, Acetivibrio thermocellus, Acetivibrio thermocellus dsm 1313
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSCRIPTION/DNA Ligands: ZN, MG |
 |
Clostridium Thermocellum Rna Polymerase Transcription Open Complex With Sigi6 And Its Promoter
Organism: Acetivibrio thermocellus dsm 1313, Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Solution Structure Of The Periplasmic Domain Of The Anti-Sigma Factor Rsgi1 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |
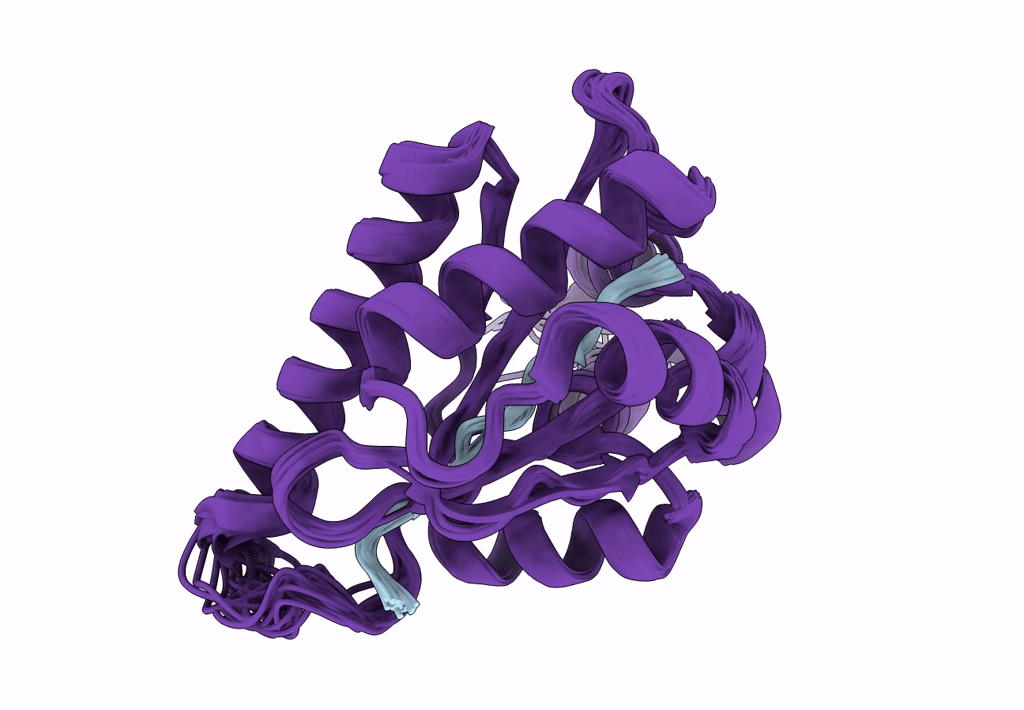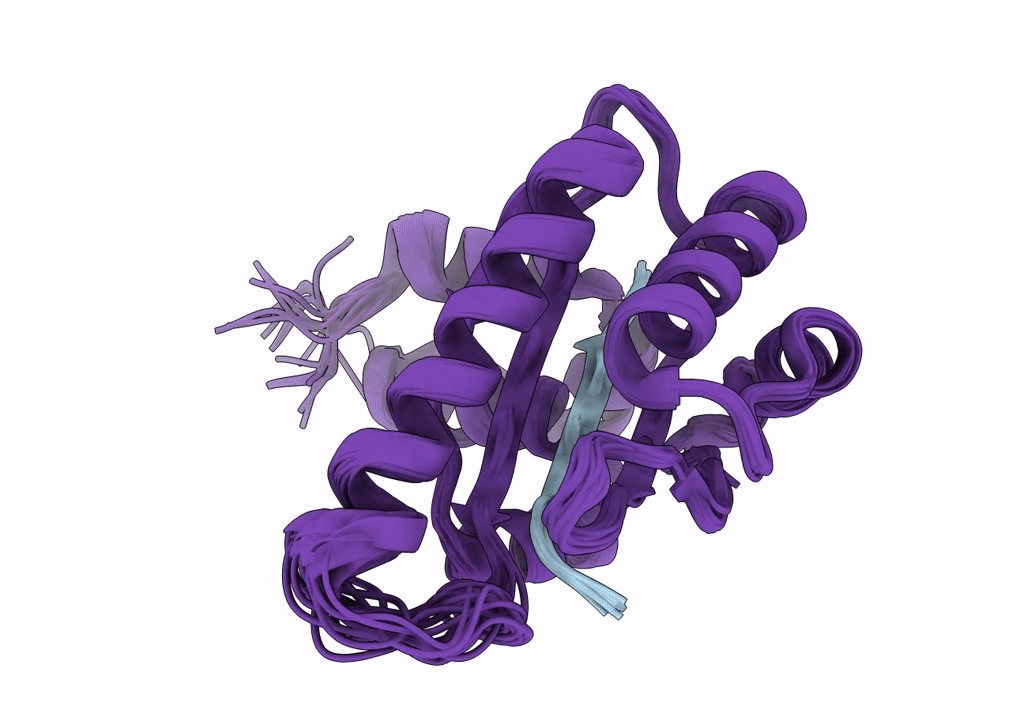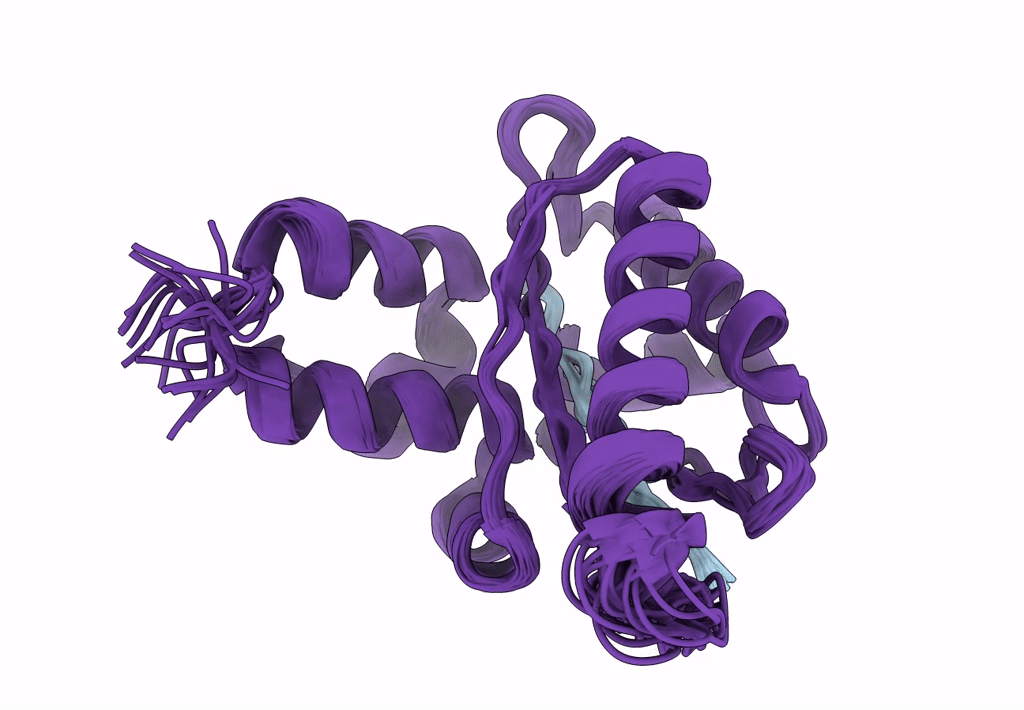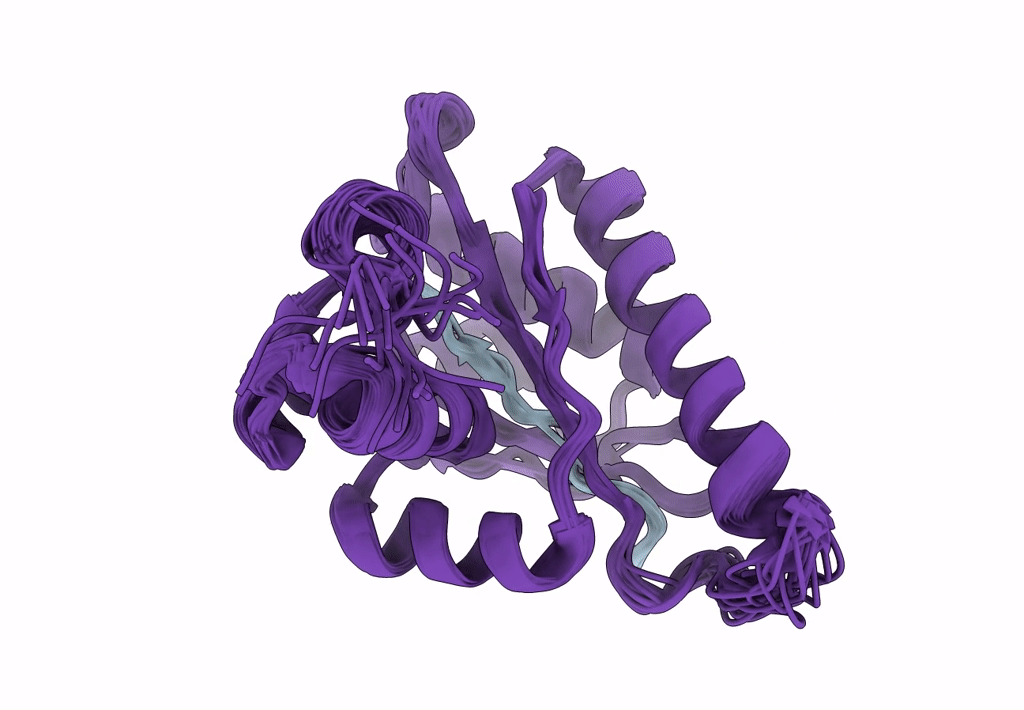 |
Solution Structure Of The Periplasmic Domain Of The Anti-Sigma Factor Rsgi2 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |
 |
Solution Structure Of The Periplasmic Domain Of Rsgi6 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |
 |
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-17 Classification: SIGNALING PROTEIN |
 |
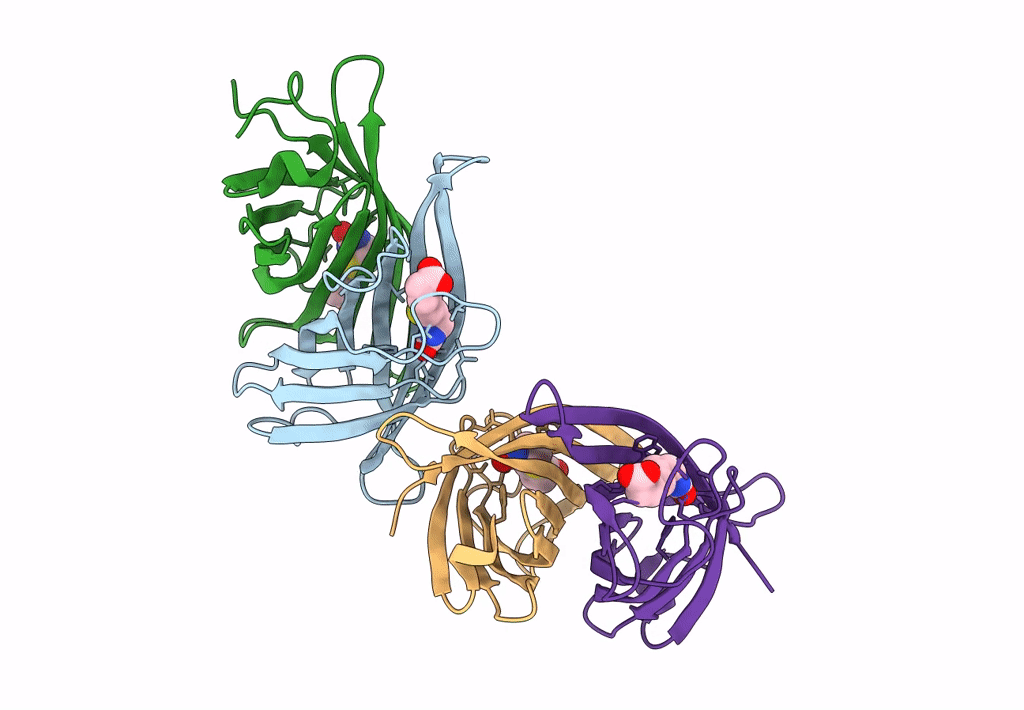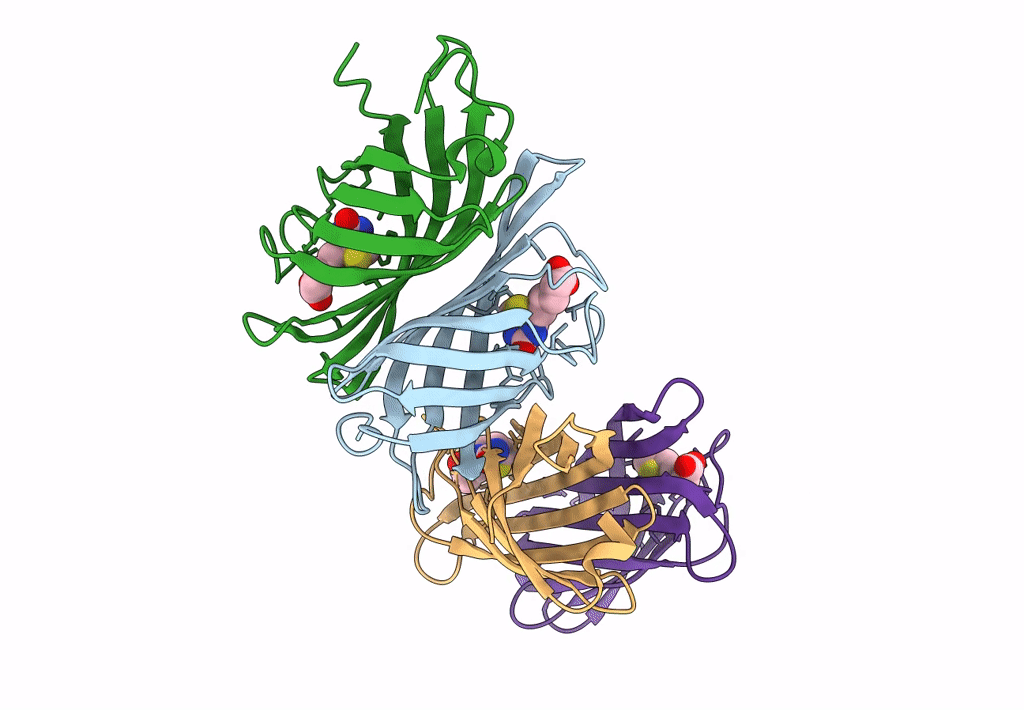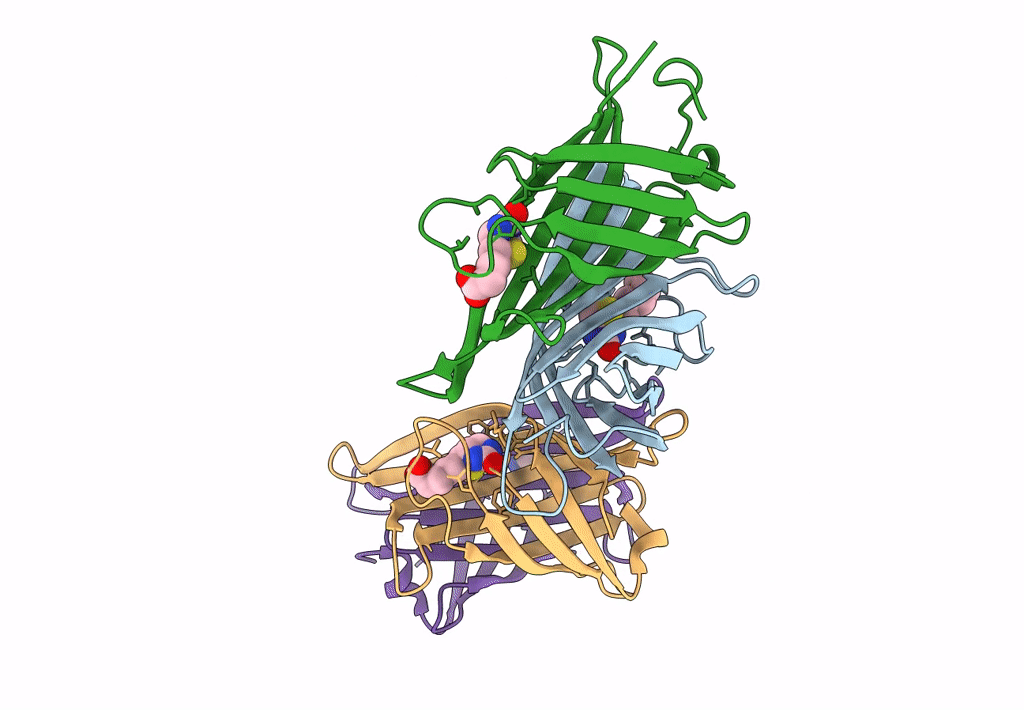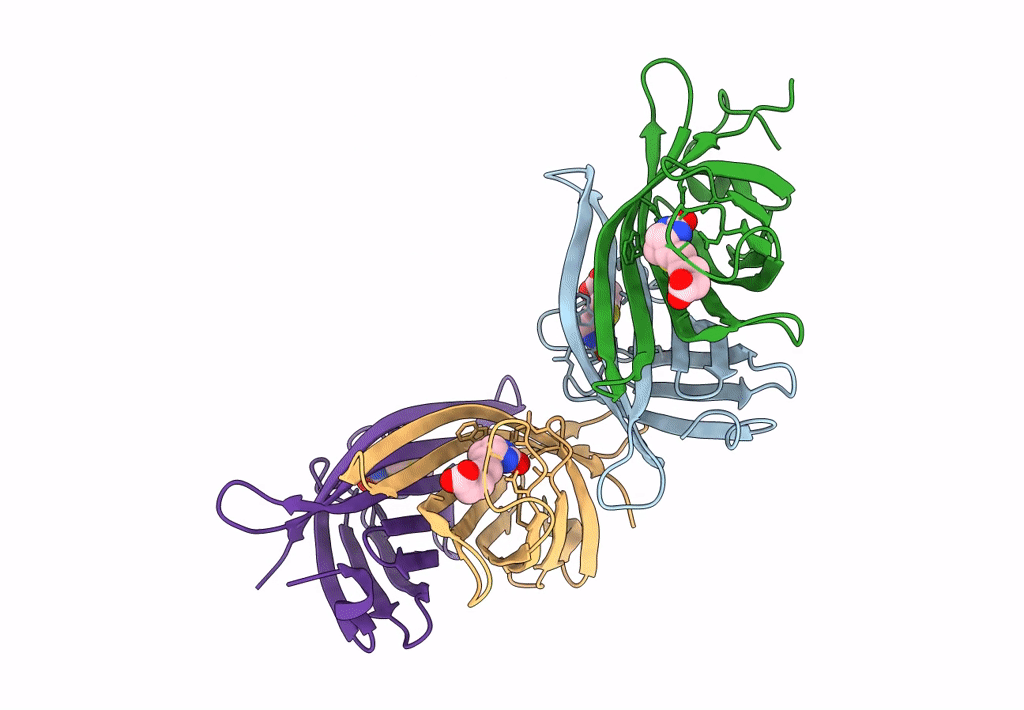
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION |
 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION Ligands: BTN |
 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION |
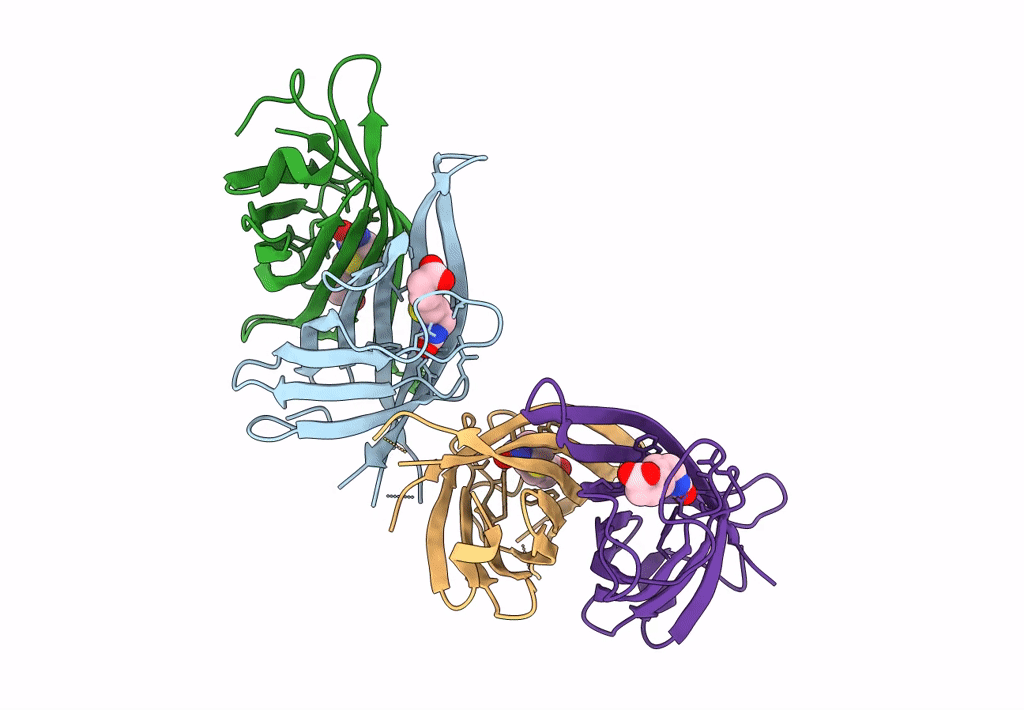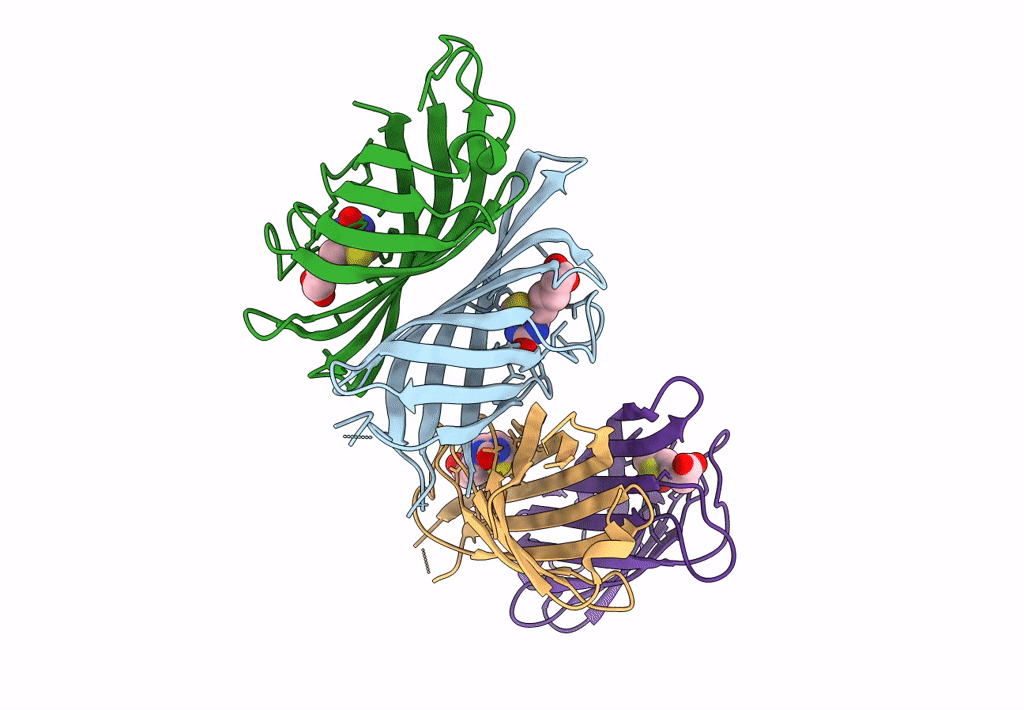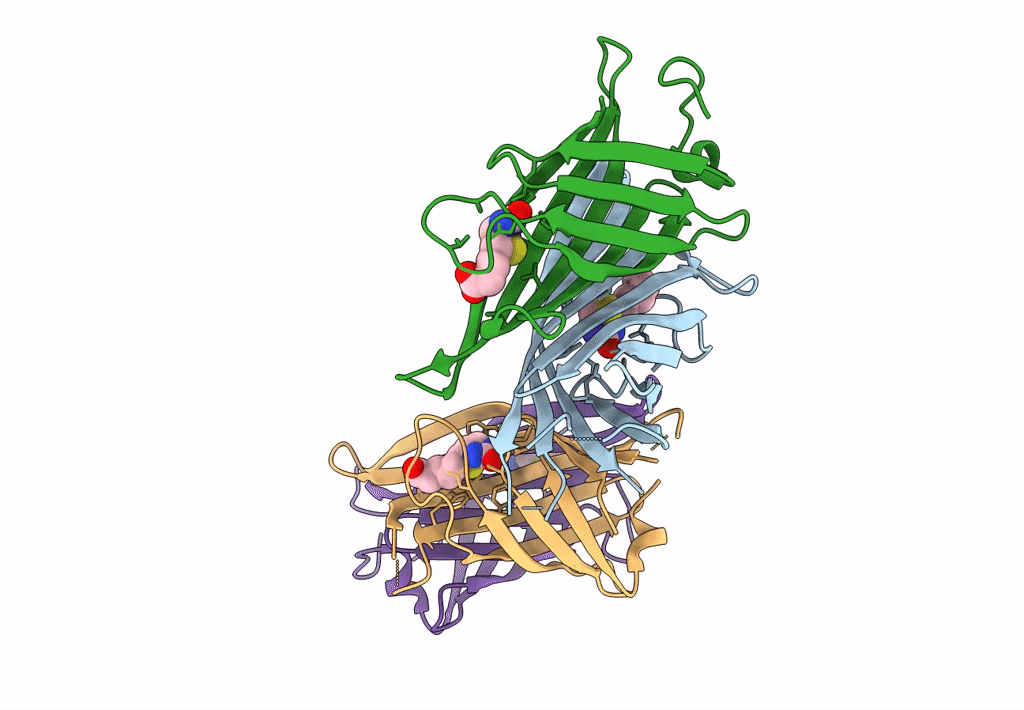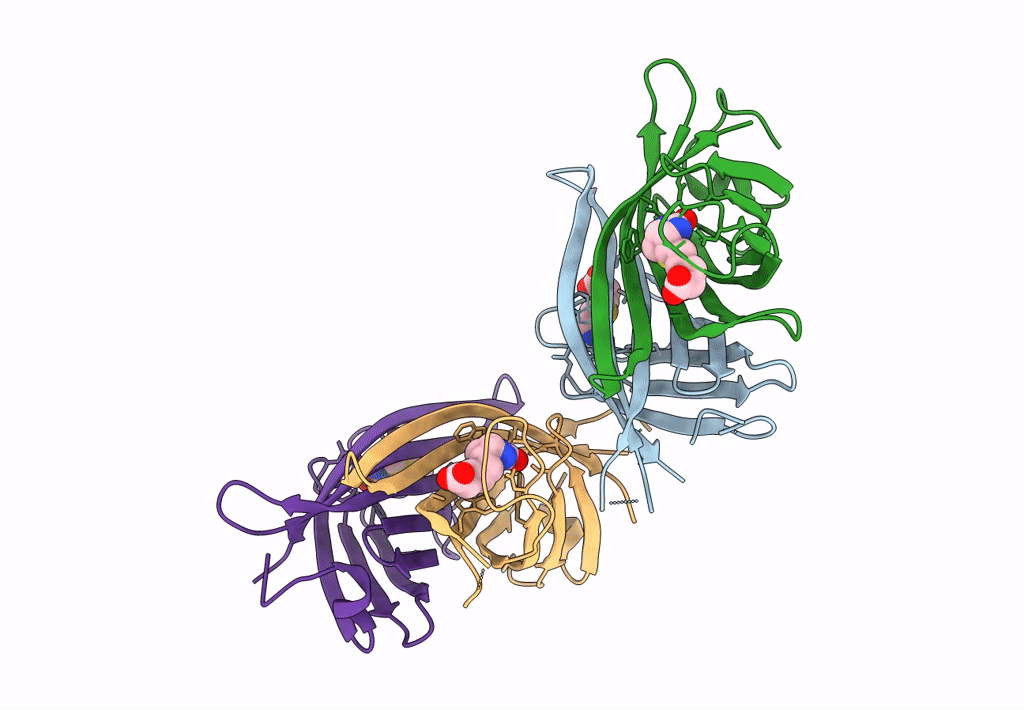 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION Ligands: BTN |
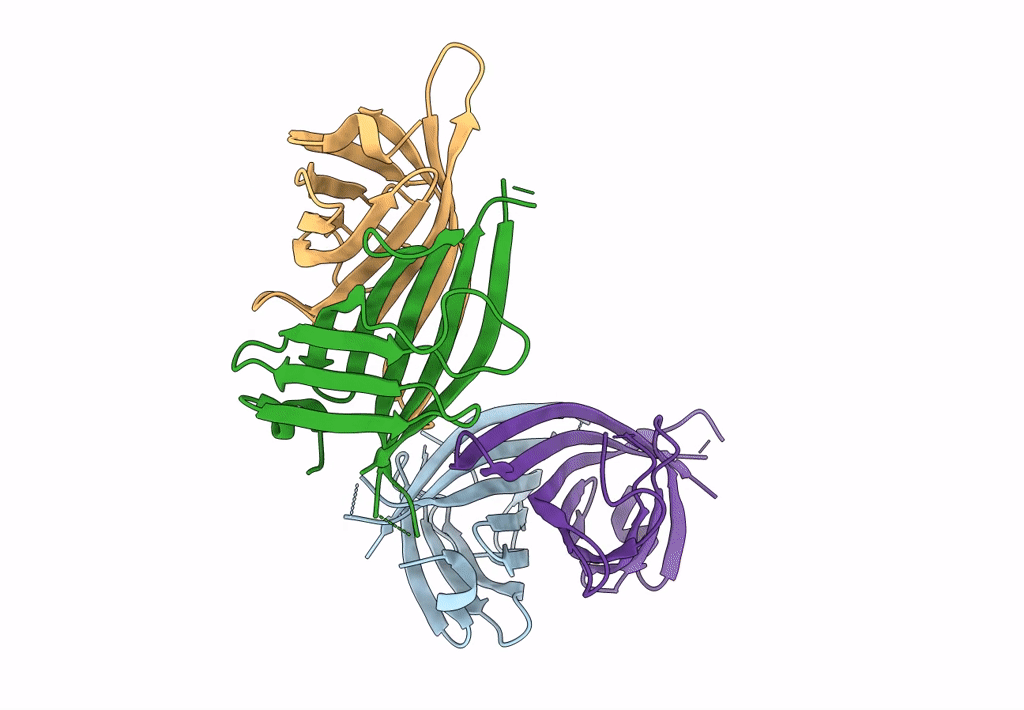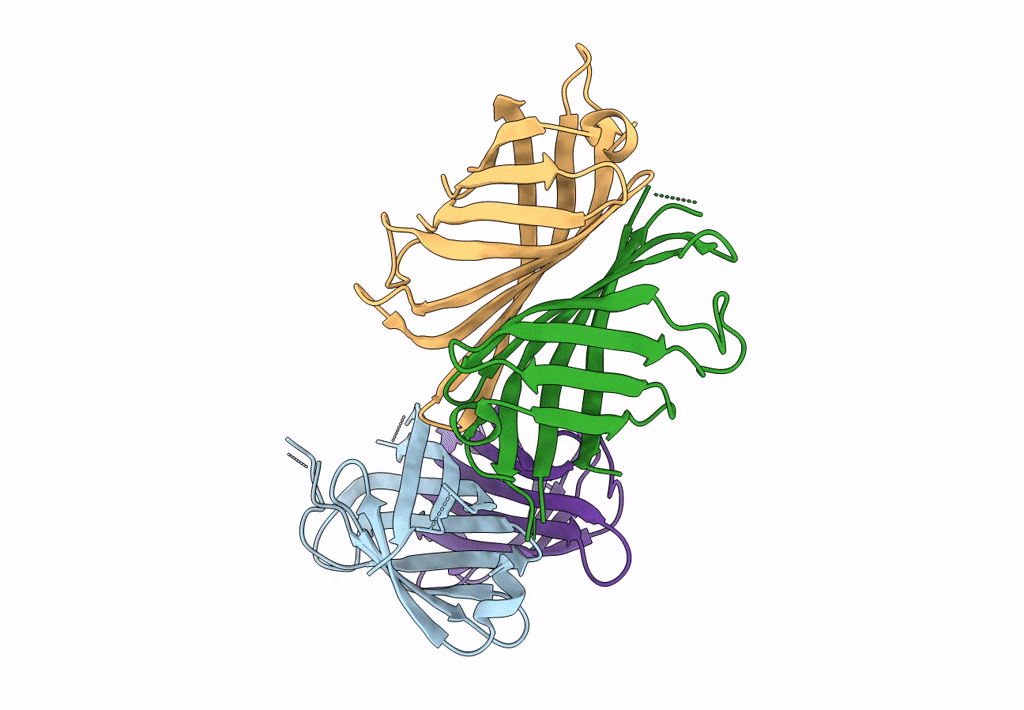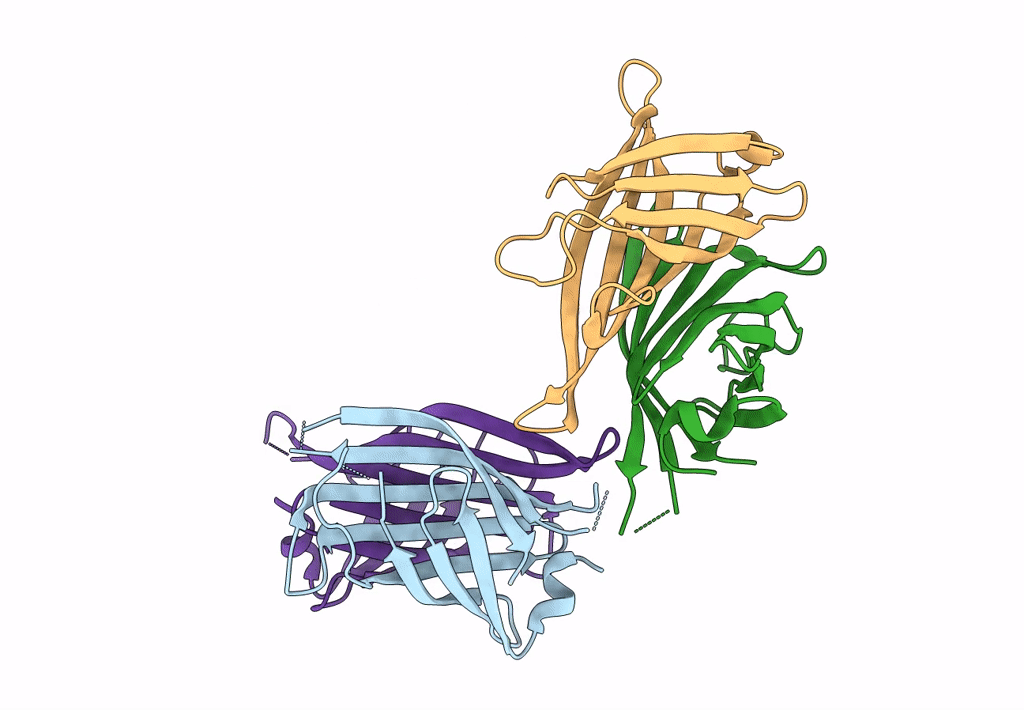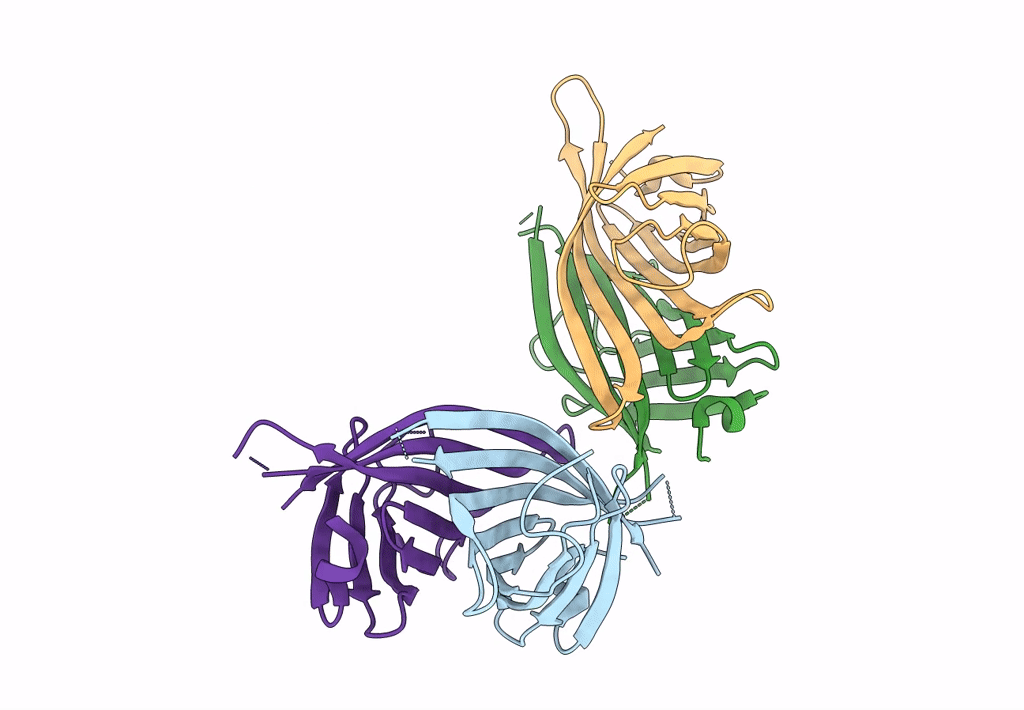 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION |
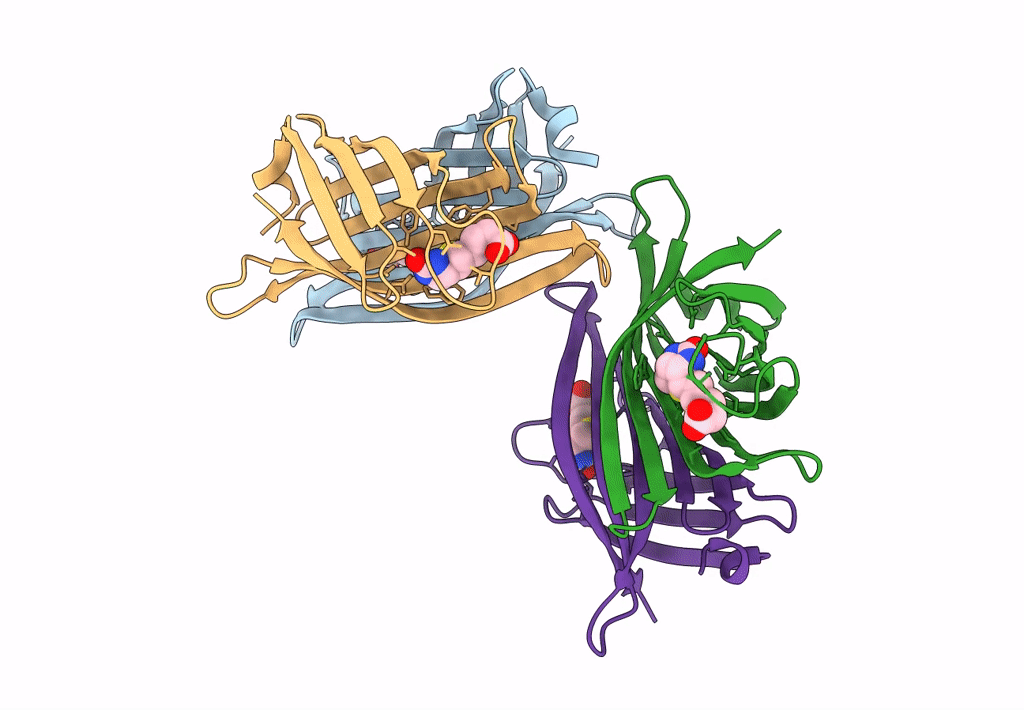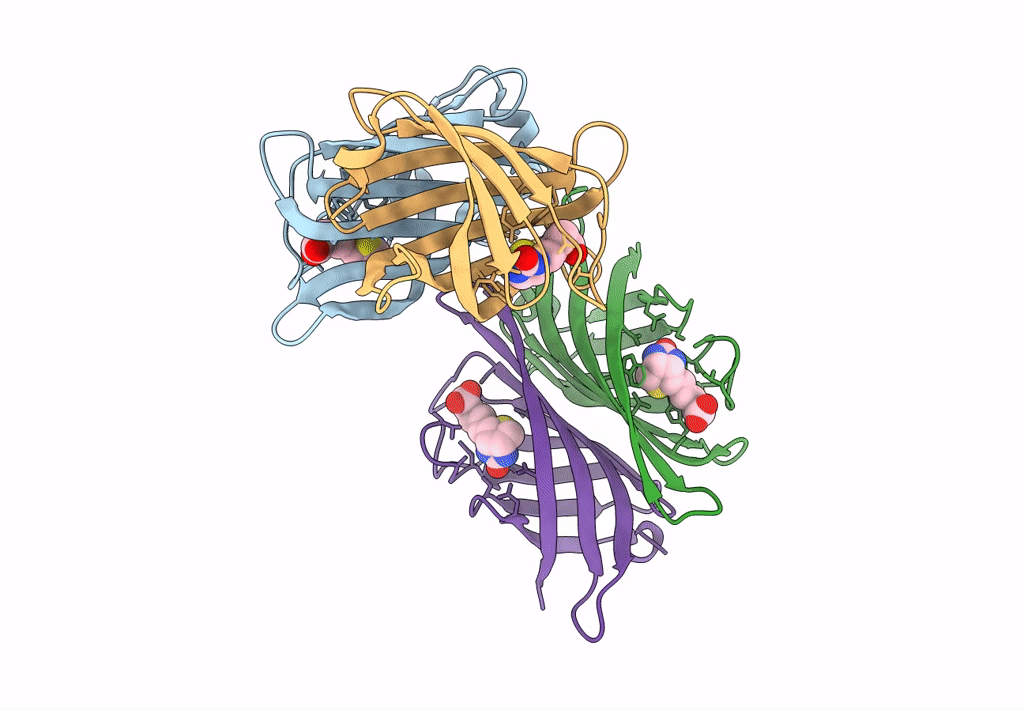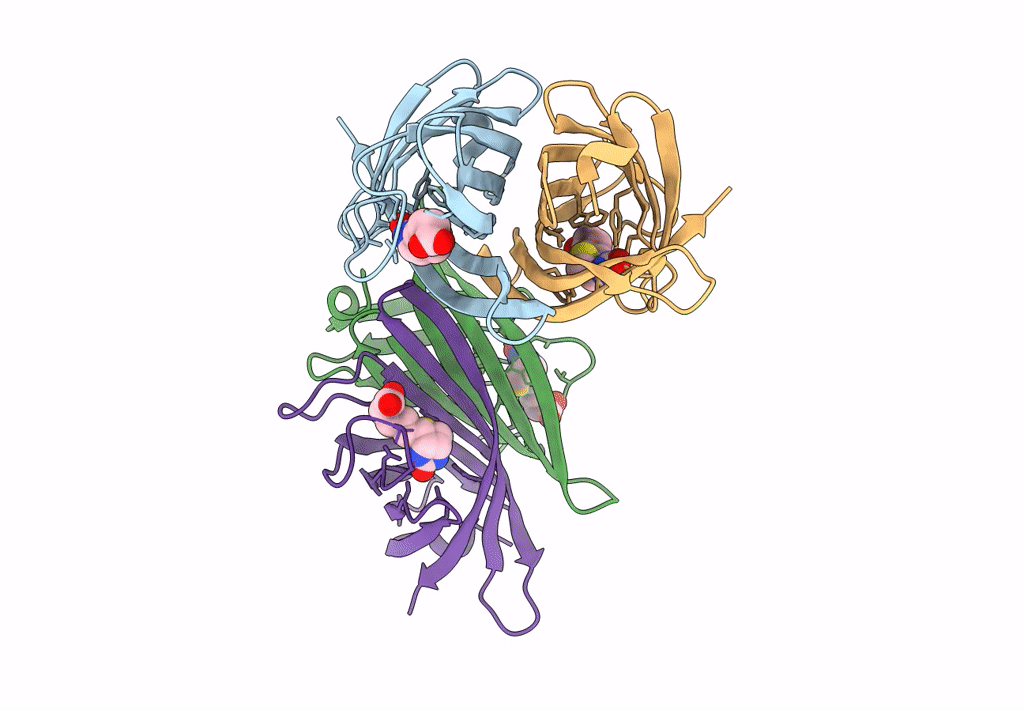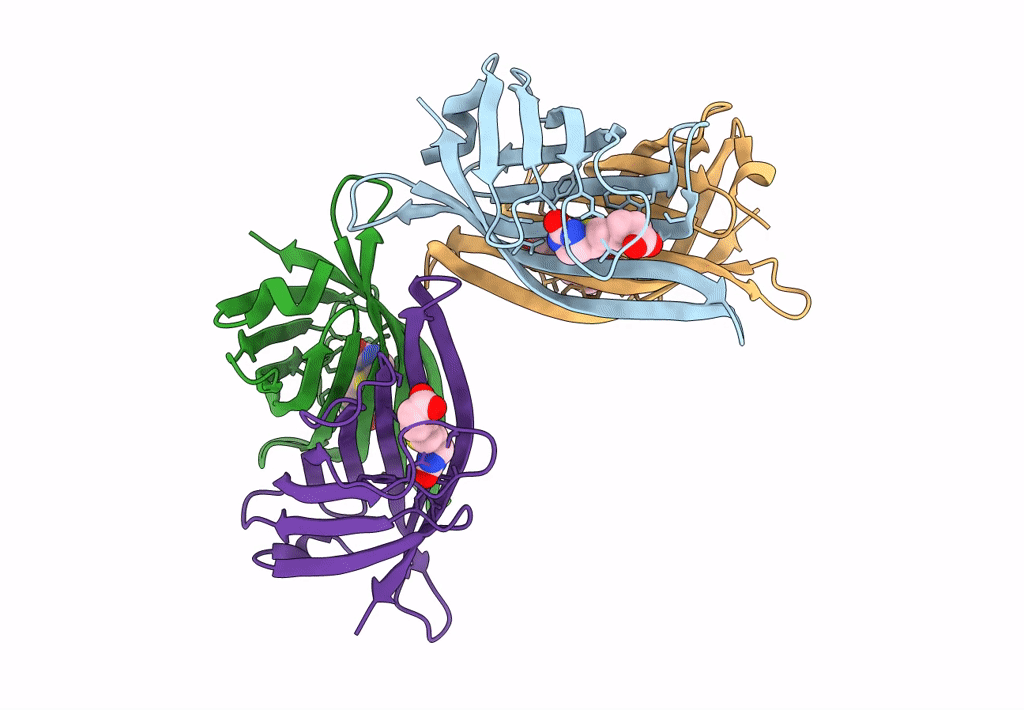 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION Ligands: BTN |
 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION Ligands: EDO |
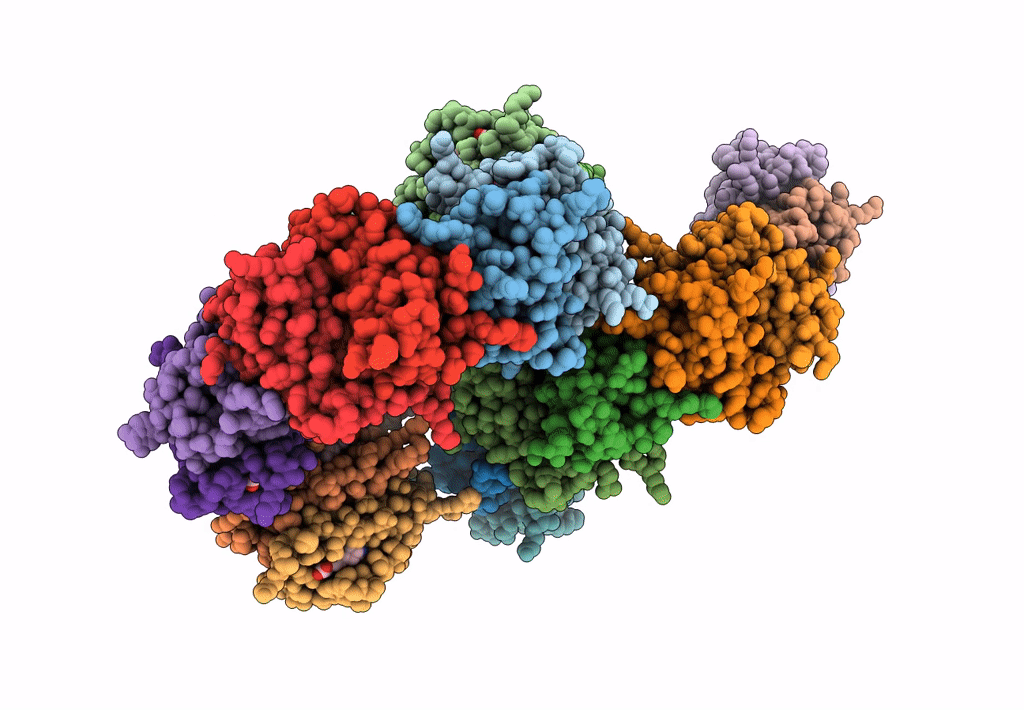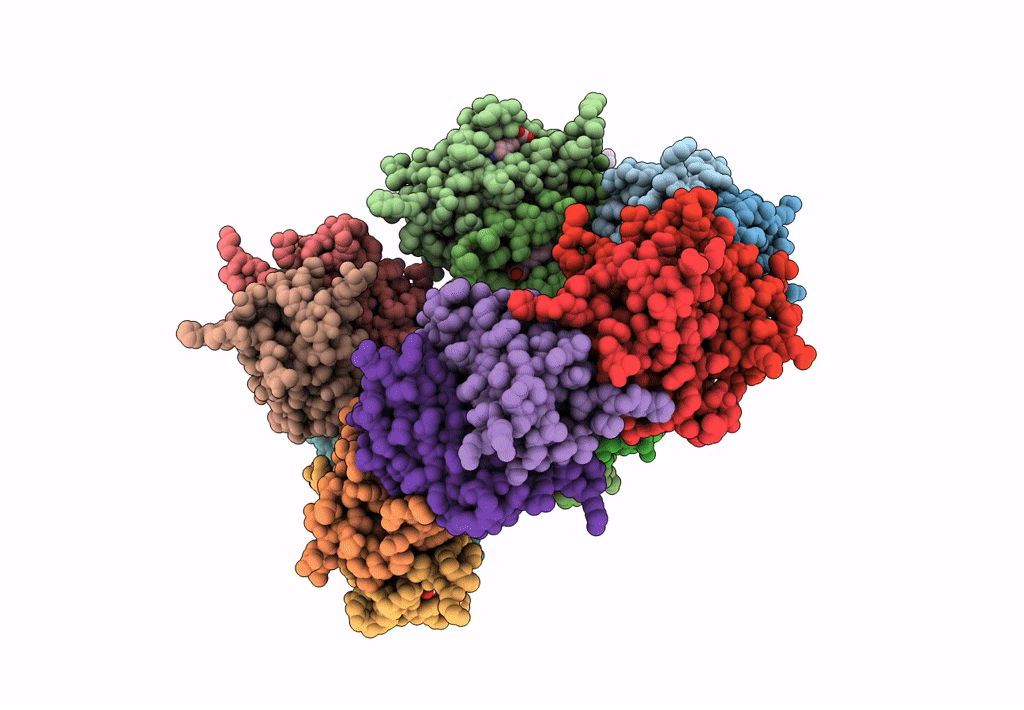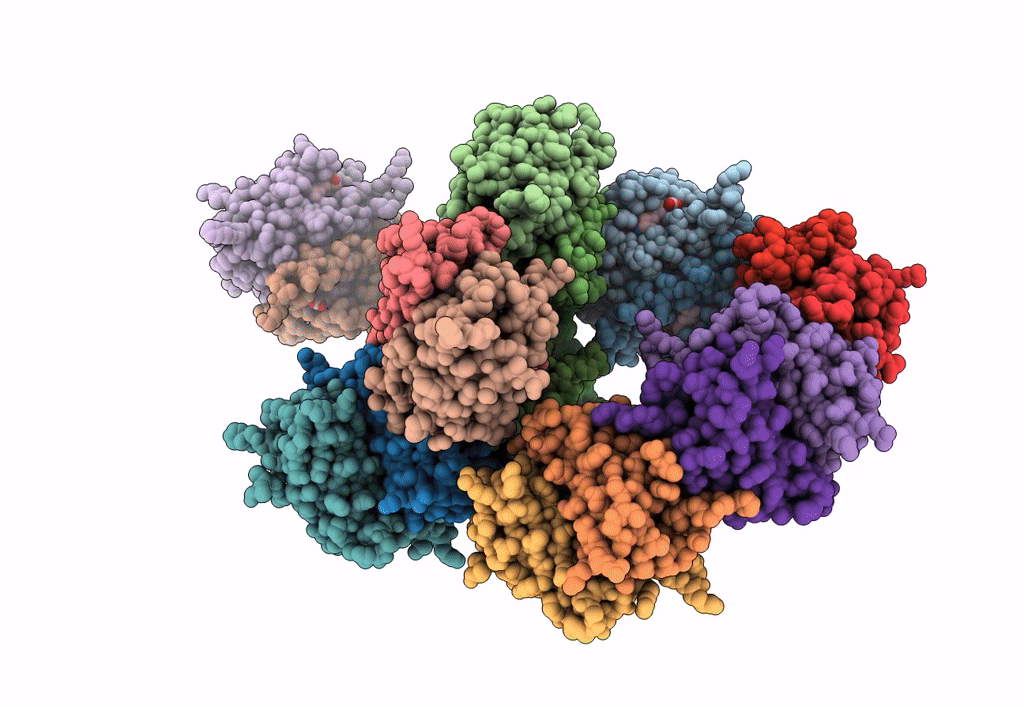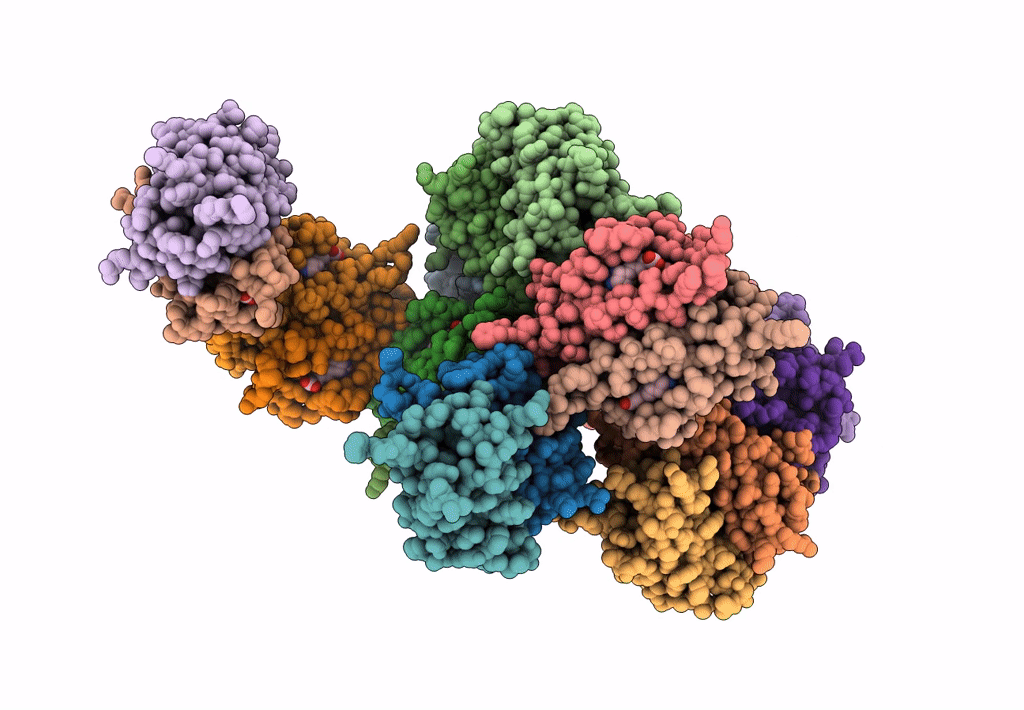 |
Organism: Rhizobium sp. aap43
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2023-04-12 Classification: UNKNOWN FUNCTION Ligands: BTN |
 |
Ruminococcus Flavefaciens Cohesin-Dockerin Structure: Dockerin From Scah Adaptor Scaffoldin In Complex With The Cohesin From Scae Anchoring Scaffoldin
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-11-02 Classification: PROTEIN BINDING Ligands: CA, GOL, SO4 |
 |
Crystal Structure Of Sugar Binding Protein Cbpa From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpa Complexed Wtih Glucose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN Ligands: BGC |
 |
Crystal Structure Of Sugar Binding Protein Cbpb From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |

