Search Count: 18
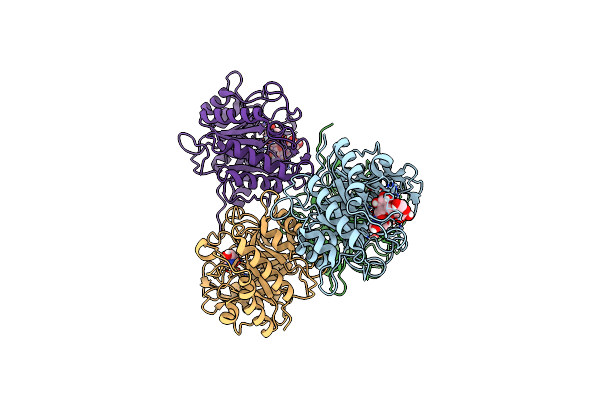 |
Crystal Structure Of Pseudomonas Stutzeri Endoglucanase Cel5A In Complex With Cellobiose
Organism: Pseudomonas stutzeri (strain a1501)
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-01-29 Classification: HYDROLASE Ligands: TRS |
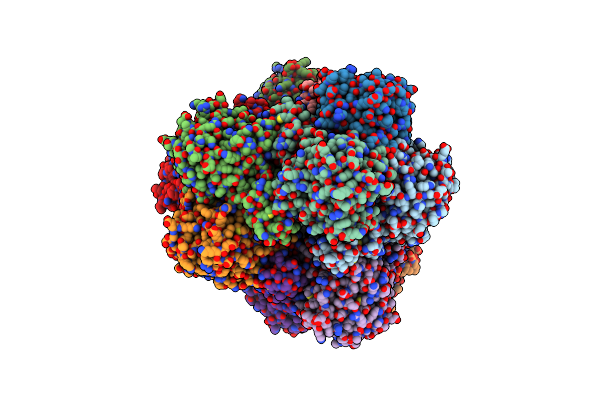 |
Crystal Structure Of The M42 Aminopeptidase Tmpep1050 From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-04-08 Classification: HYDROLASE |
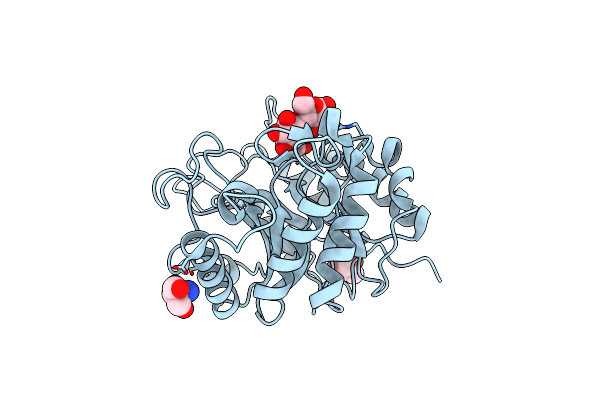 |
Crystal Structure Of The Endo-1,4-Glucanase, Rbcel1, In Complex With Cellobiose
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2014-12-31 Classification: HYDROLASE Ligands: TRS |
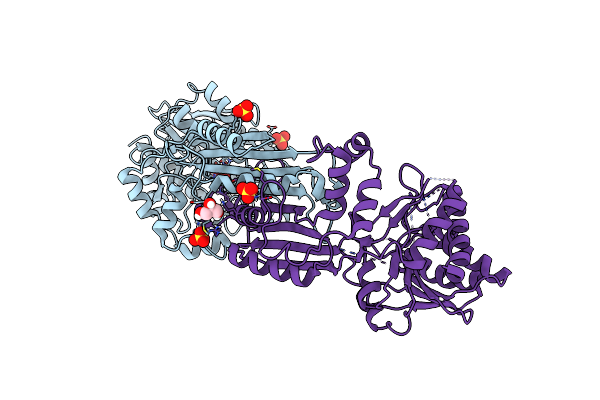 |
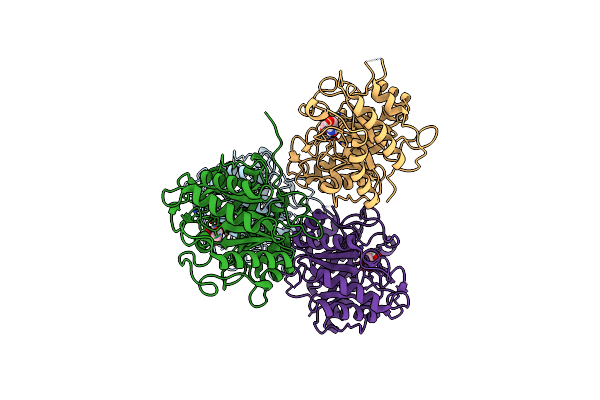
The Crystal Structure Of A M20 Family Metallo-Carboxypeptidase Sso-Cp2 From Sulfolobus Solfataricus
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2014-10-15 Classification: HYDROLASE Ligands: ZN, SO4, GOL |
 |
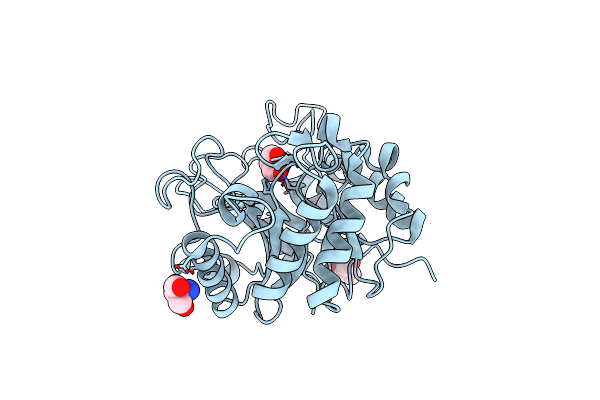
Crystal Structure Determination Of Pseudomonas Stutzeri Endoglucanase Cel5A Using A Twinned Data Set
Organism: Pseudomonas stutzeri
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2014-07-30 Classification: HYDROLASE Ligands: TRS |
 |
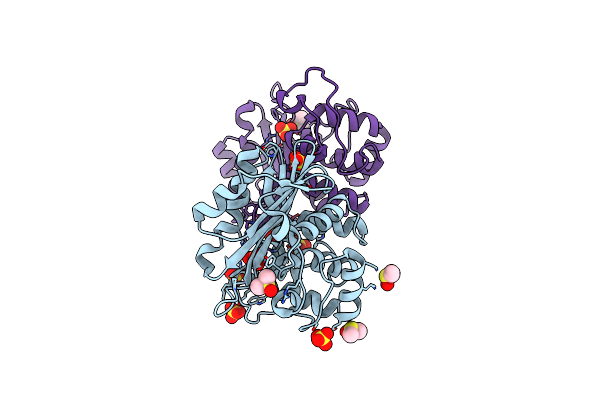
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2013-04-03 Classification: HYDROLASE Ligands: TRS |
 |
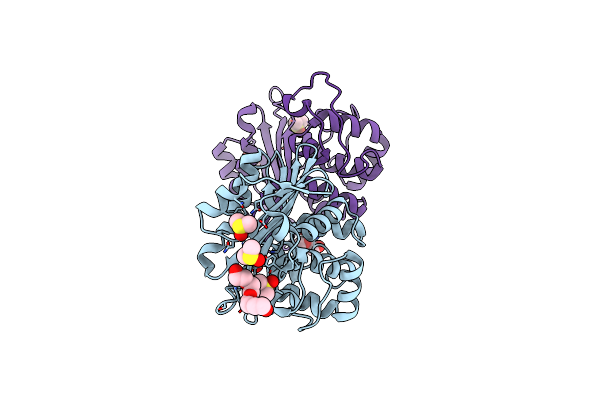
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-12-08 Classification: HYDROLASE Ligands: SO4, DMS, PG4 |
 |
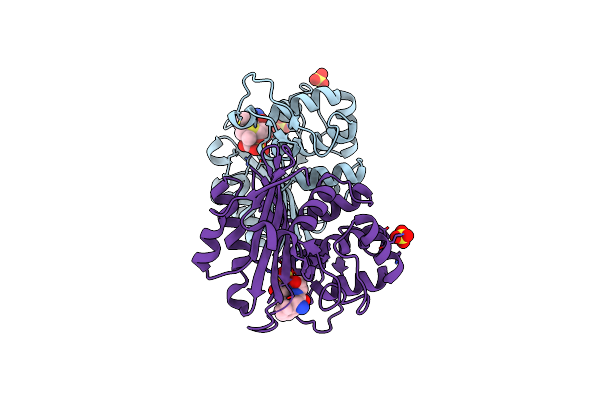
Crystal Structure Of The Class D Beta-Lactamase Oxa-10 At 1.35 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2010-12-08 Classification: HYDROLASE Ligands: SO4, PG4, DMS, GOL |
 |
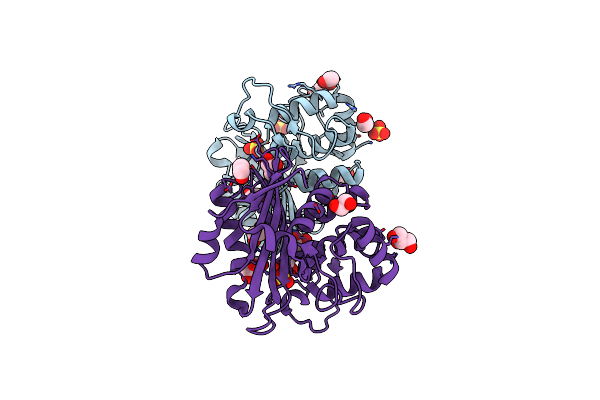
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: ZZ7, SO4 |
 |
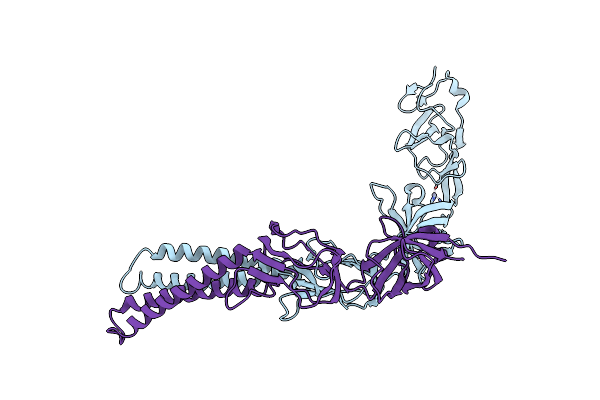
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: SO4, GOL, EDO |
 |
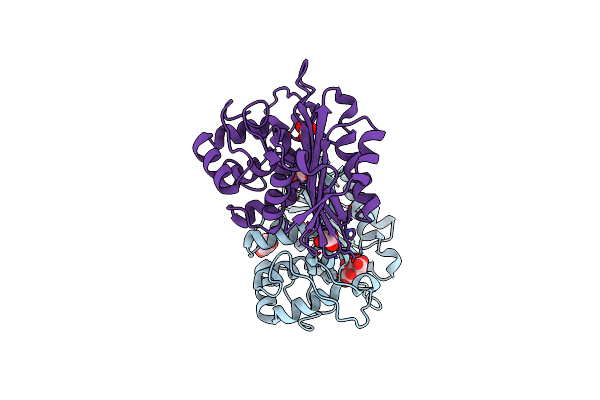
Organism: Cupriavidus metallidurans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-06-30 Classification: METAL TRANSPORT Ligands: ZN |
 |
Crystal Structure Of The Oxa-10 V117T Mutant At Ph 6.5 Inhibited By A Chloride Ion
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: GOL, CL, CIT, SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-12-08 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-23 Classification: HYDROLASE |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-08-05 Classification: METAL BINDING PROTEIN |
 |
Organism: Colwellia psychrerythraea
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-07-01 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Plasmid-Encoded Class C Beta-Lactamase Cmy-2 Complexed With Citrate Molecule
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2006-04-25 Classification: HYDROLASE Ligands: CIT |

