Search Count: 179
 |
Structure Of The Pro-Pro Endopeptidase (Ppep-3) E153A Y189F In Complex With Substrate Peptide Ac-Eplpppp-Nh2 From Geobacillus Thermodenitrificans
Organism: Geobacillus thermodenitrificans ng80-2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: ZN, PG4, PGE |
 |
Structure Of The Pro-Pro Endopeptidase (Ppep-3) E153A Y189F From Geobacillus Thermodenitrificans
Organism: Geobacillus thermodenitrificans ng80-2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: ZN, BCN, PG4, PEG |
 |
Structure Of The Pro-Pro Endopeptidase (Ppep-3) From Geobacillus Thermodenitrificans
Organism: Geobacillus thermodenitrificans ng80-2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ZN, BCN, PEG, EDO |
 |
Organism: Burkholderia pyrrocinia, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Parachlamydia sp., Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: TLA |
 |
Organism: Homo sapiens, Parachlamydia sp.
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Pigmentiphaga aceris, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: AYE |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-12-13 Classification: HYDROLASE |
 |
Tssm-Ub-Pa Complex - A Usp-Like Dub From B. Pseudomallei (193-430) Reacted With Ub-Pa
Organism: Burkholderia pseudomallei, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: FLC, EDO, NA |
 |
Linear Specific Otu-Type Dub Snotu From The Pathogen S. Negenvensis In Complex With Linear Di-Ubiquitin
Organism: Simkania negevensis z, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-11-22 Classification: HYDROLASE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2023-11-08 Classification: HYDROLASE Ligands: APR |
 |
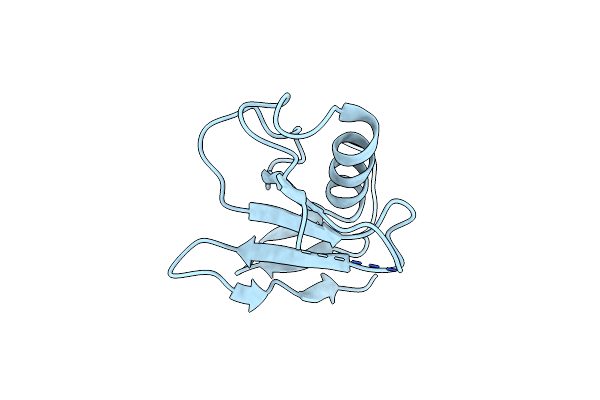
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION Ligands: HOH |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2(1)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Hordei Avra22
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Hordei Avra10
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
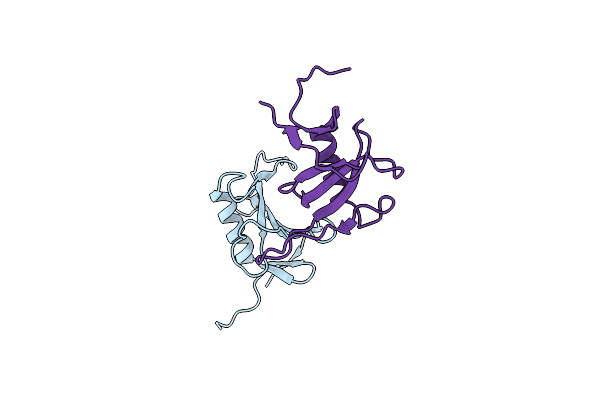
Organism: Blumeria hordei dh14
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2 (2)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Organism: Waddlia chondrophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: CIT, AYE |
 |
Organism: Waddlia chondrophila
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-15 Classification: HYDROLASE |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-15 Classification: HYDROLASE |

