Search Count: 18
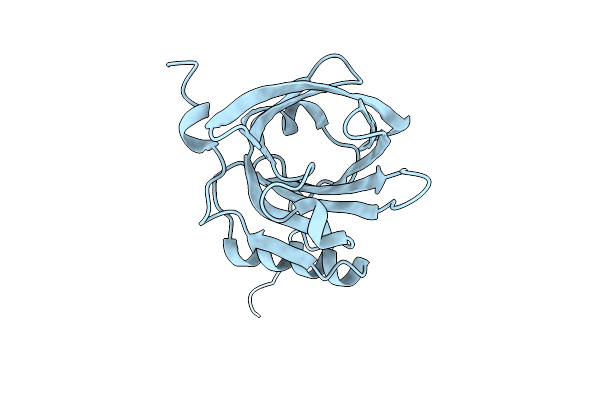 |
Organism: Felis catus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-12-03 Classification: ALLERGEN |
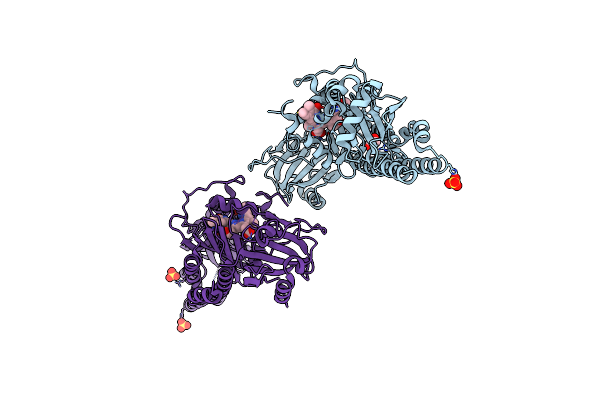 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
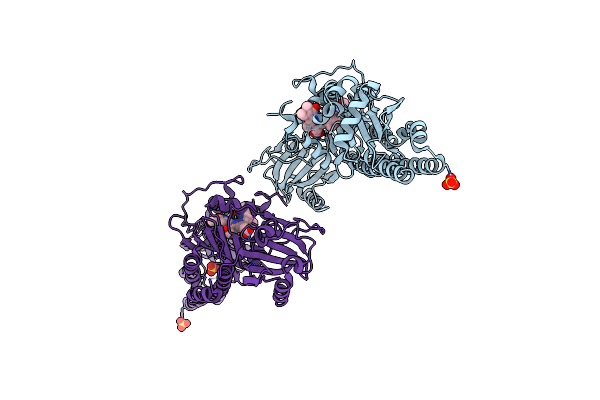 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
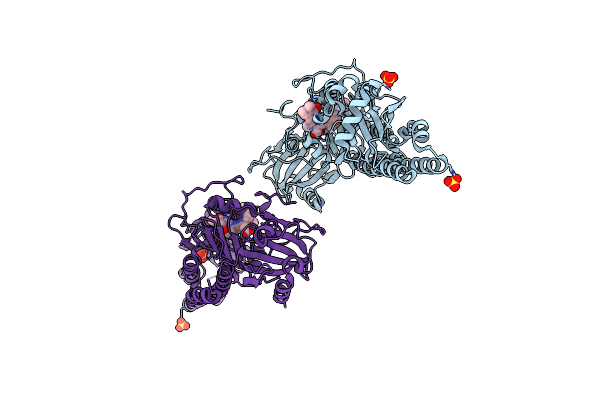 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Felis catus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-08-16 Classification: ALLERGEN Ligands: SO4 |
 |
Cryo-Em Structure Of The Agonist Setmelanotide Bound To The Active Melanocortin-4 Receptor (Mc4R) In Complex With The Heterotrimeric Gs Protein At 2.6 A Resolution.
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Active Melanocortin-4 Receptor (Mc4R)- Gs Protein Complex Bound To Agonist Ndp-Alpha-Msh At 2.86 A Resolution.
Organism: Homo sapiens, Rattus norvegicus, Mus musculus, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Conformational Changes Of The Clamp Of The Protein Translocation Atpase Seca From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-06-03 Classification: PROTEIN TRANSPORT Ligands: ADP, MG |
 |
Improved Variant Of (R)-Selective Manganese-Dependent Hydroxynitrile Lyase From Bacteria
Organism: Granulicella tundricola
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-01-21 Classification: LYASE Ligands: MN |
 |
Structure Determination Of The Ras-Binding Domain Of The Ral-Specific Guanine Nucleotide Exchange Factor Rlf, Nmr, 10 Structures
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 1999-02-16 Classification: SIGNAL TRANSDUCTION PROTEIN |
 |
Structure Determination Of Antiviral Compound Sch 38057 Complexed With Human Rhinovirus 14
Organism: Human rhinovirus 14
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1993-10-31 Classification: VIRUS Ligands: S57 |

