Search Count: 226
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Co-Crystal Structure Of Yeast Forkhead Transcription Factor Fkh1 Bound To Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: DNA BINDING PROTEIN/DNA Ligands: K |
 |
Chloride Bound Structure Of Oxidized Ba3-Type Cytochrome C Oxidase Confirmed By Single-Wavelength Anomalous Diffraction
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HAS, OLC, CL, CUA |
 |
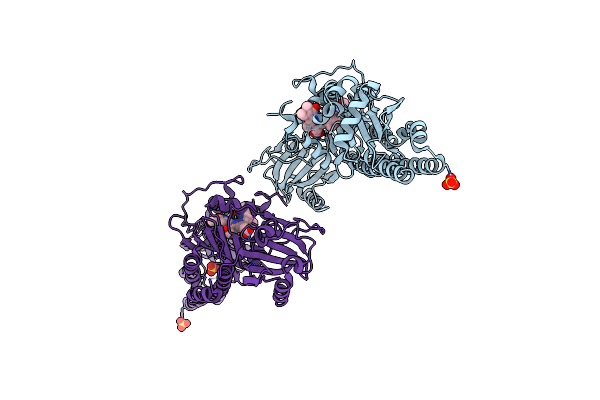
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
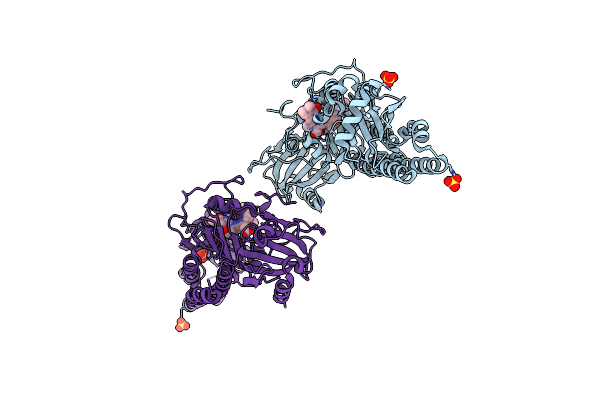
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
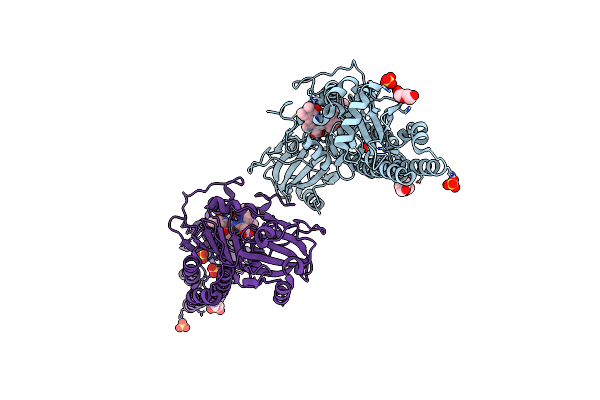
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
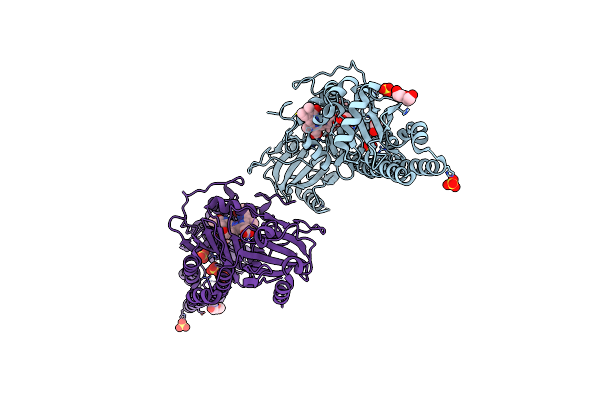
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Time-Resolved Sfx-Xfel Crystal Structure Of Cyp121 Bound With Cyy Reacted With Peracetic Acid For 200 Milliseconds
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT, PEO |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Catalytically Active Radical State Solved By Xfel
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Radical-Lost Ground State
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: CA, GOL |

