Search Count: 42
 |
Organism: Streptococcus canis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Structure Of The Truncated Version Of Idec Protease C94S From Streptococcus Canis
Organism: Streptococcus canis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
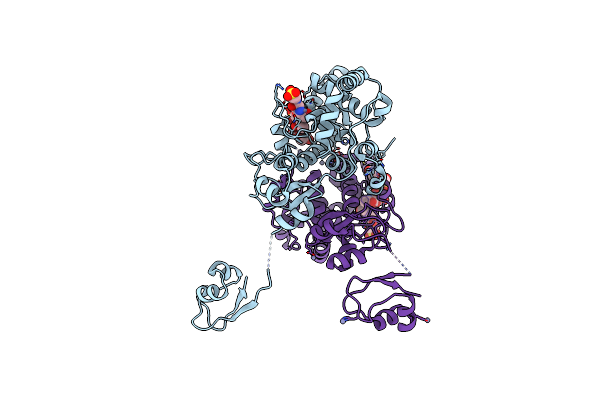
Lytic Transglycosylase Mltd Of Pseudomonas Aeruginosa Bound To The Natural Product Bulgecin A
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-17 Classification: LYASE Ligands: BLG, ZN |
 |
Lytic Transglycosylase Mltd Of Pseudomonas Aeruginosa Bound To The Natural Product Bulgecin A, With Two Lysm Domains
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-17 Classification: LYASE Ligands: PEG, BLG, ZN |
 |
Lytic Transglycosylase Mltd Of Pseudomonas Aeruginosa In A Ternary Complex Bound To Bulgecin A And Chito-Tetraose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-04-17 Classification: LYASE Ligands: BLG, ZN |
 |
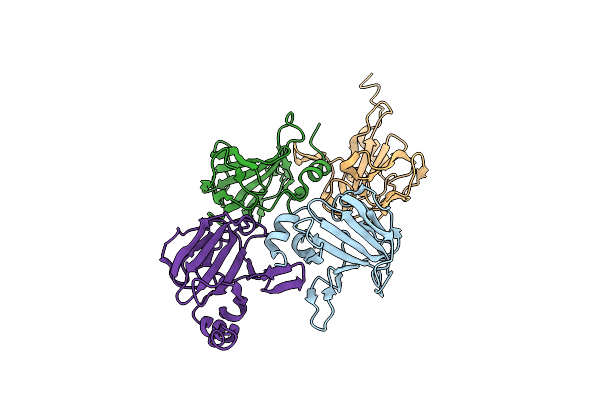
X-Ray Structure Of The Lytic Transglycosylase Sltb2 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: CA |
 |
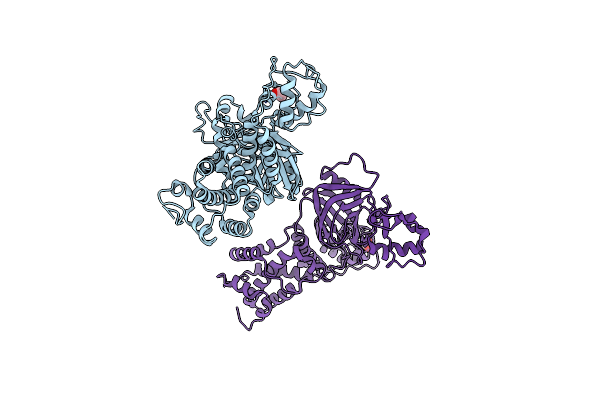
Lytm Domain Of Dipm, A Coordinator Of A Complex Net Of Autolysins In Caulobacter Crescentus
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-06-21 Classification: CELL CYCLE |
 |
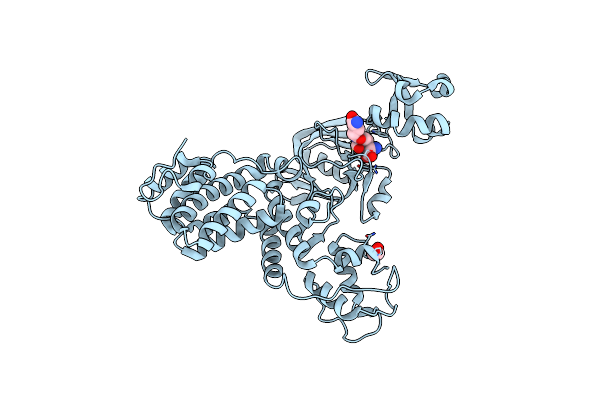
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2021-10-06 Classification: TRANSFERASE Ligands: EDO |
 |
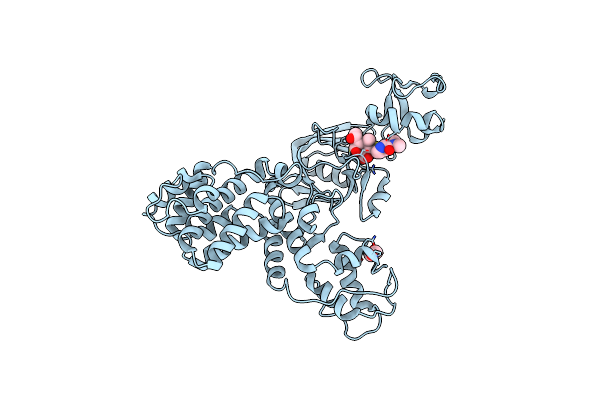
The X-Ray Structure Of L,D-Transpeptidase Ldta From Vibrio Cholerae In Complex With The Cross-Linking Reaction Intermediate
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-10-06 Classification: TRANSFERASE Ligands: EDO, S2K |
 |
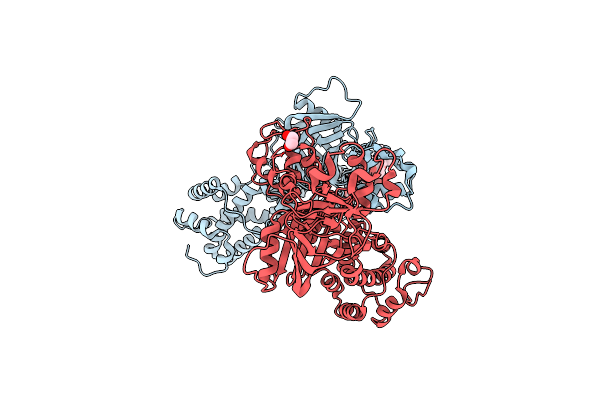
The X-Ray Structure Of L,D-Transpeptidase Ldta From Vibrio Cholerae In Complex With Meropenem
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-10-06 Classification: TRANSFERASE Ligands: MXR, EDO |
 |
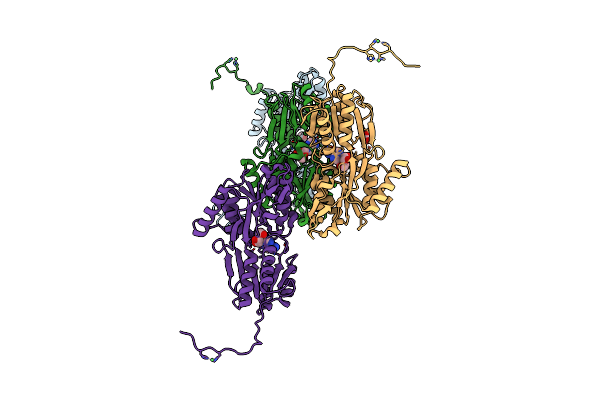
The X-Ray Structure Of L,D-Transpeptidase Ldta From Vibrio Cholerae In Complex With Nag-Nam(Tetrapeptide)
Organism: Vibrio cholerae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2021-10-06 Classification: TRANSFERASE Ligands: EDO, AMV |
 |
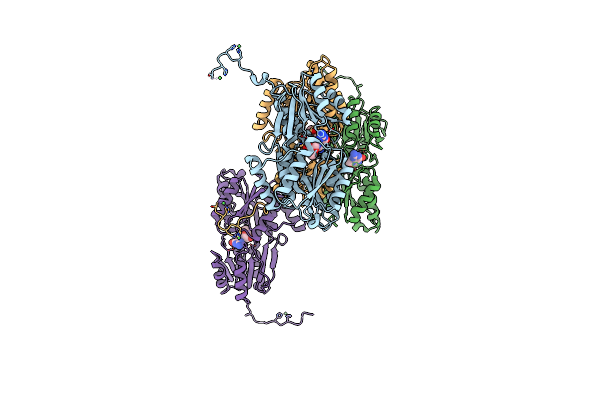
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-02-10 Classification: TRANSPORT PROTEIN Ligands: NI, ADN, ACT |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-02-10 Classification: TRANSPORT PROTEIN Ligands: GMP, NI |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-02-10 Classification: TRANSPORT PROTEIN Ligands: CTN, NI, ACT, PEG |
 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-10 Classification: TRANSPORT PROTEIN Ligands: NI, URI, CAC, ACT |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-02-10 Classification: TRANSPORT PROTEIN Ligands: NI, THM, ACT |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx From P. Aeruginosa In Complex With 1-Deoxynojirimycin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, NOJ, MG |
 |
The Crystal Structure The Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Two Glucose Molecules
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Glucose
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |

