Search Count: 19
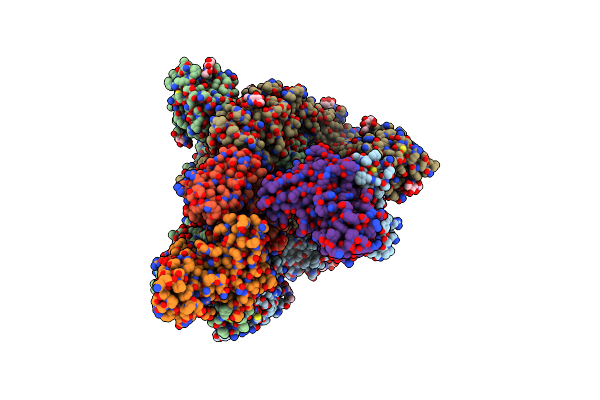 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: VIRAL PROTEIN Ligands: NAG |
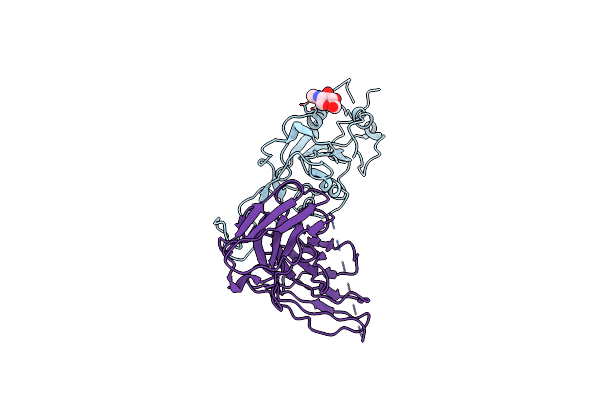 |
Sars-Cov-2 Spike Rbd In Complex With The Single Chain Fragment Scfv76 (Focused Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: VIRAL PROTEIN Ligands: NAG |
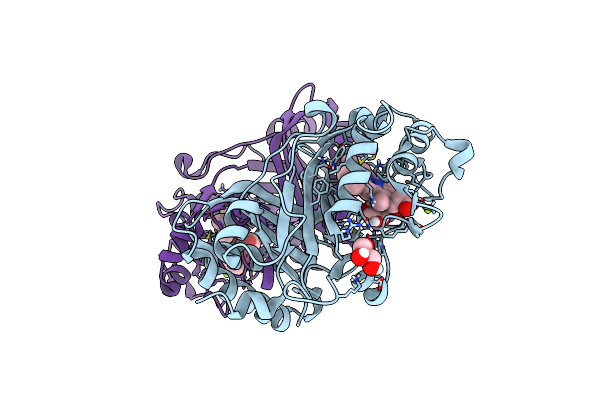 |
Crystal Structure Of A Dye-Decolorizing Peroxidase D143Ar232A Variant From Klebsiella Pneumoniae (Kpdyp)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, MG |
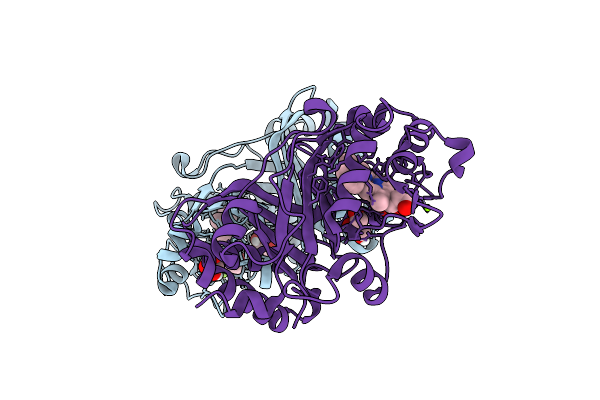 |
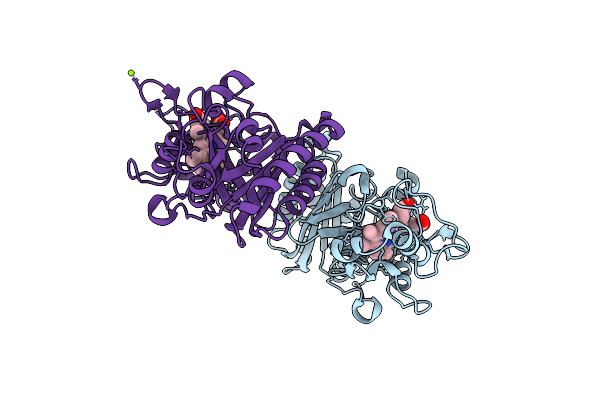
Crystal Structure Of A Dye-Decolorizing Peroxidase From Klebsiella Pneumoniae (Kpdyp)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, MG |
 |
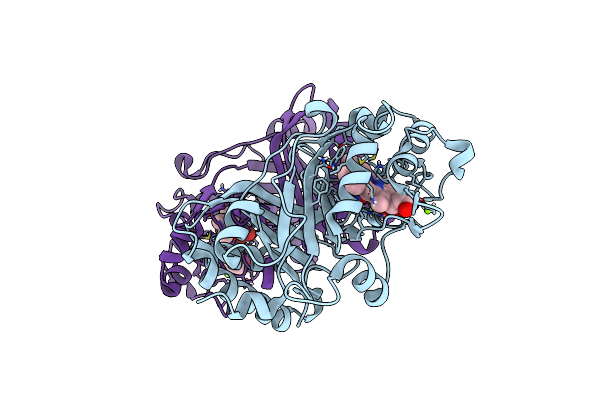
Crystal Structure Of A Dye-Decolorizing Peroxidase R232A Variant From Klebsiella Pneumoniae (Kpdyp)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: HEM, MG |
 |
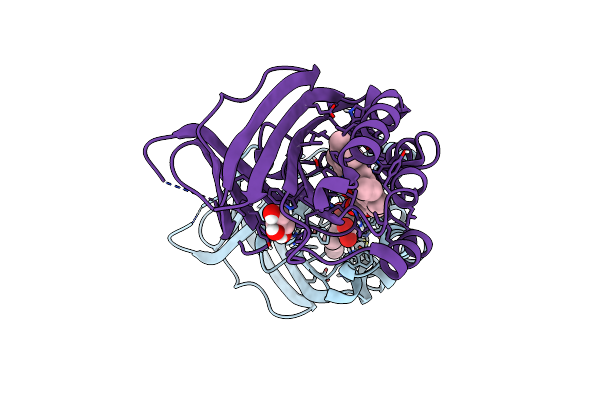
Crystal Structure Of A Dye-Decolorizing Peroxidase D143A Variant From Klebsiella Pneumoniae (Kpdyp)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: HEM, MG, NO2, GOL |
 |
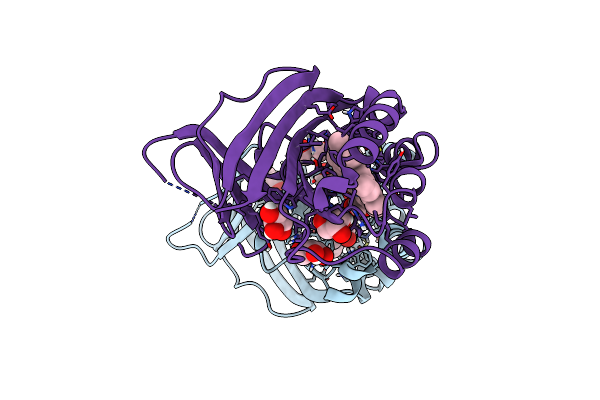
Joint Neutron/X-Ray Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.000 Å, 2.350 Å Release Date: 2018-02-28 Classification: OXIDOREDUCTASE Ligands: HEM, OH, CL, GOL |
 |
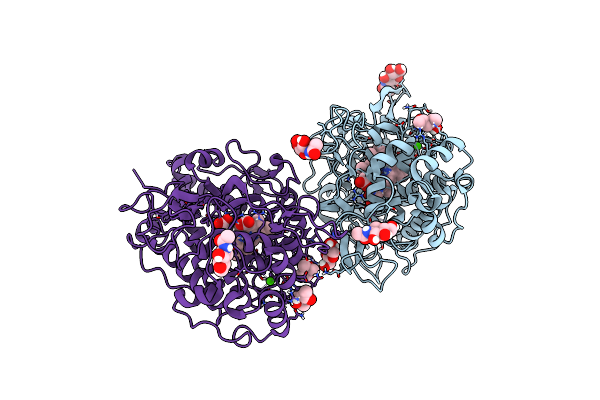
Crystal Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425 At Ph 9.0 And 293 K.
Organism: Cyanothece sp. pcc 7425
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-01-31 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, CL |
 |
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-12-27 Classification: OXIDOREDUCTASE Ligands: NAG, HEM, CA, BMA, MPD, CL, NA, MRD |
 |
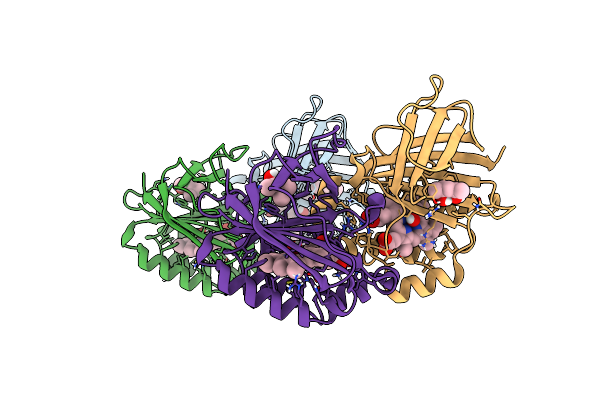
Crystal Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425 (Ph 6.5)
Organism: Cyanothece sp. pcc 7425
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, GOL |
 |
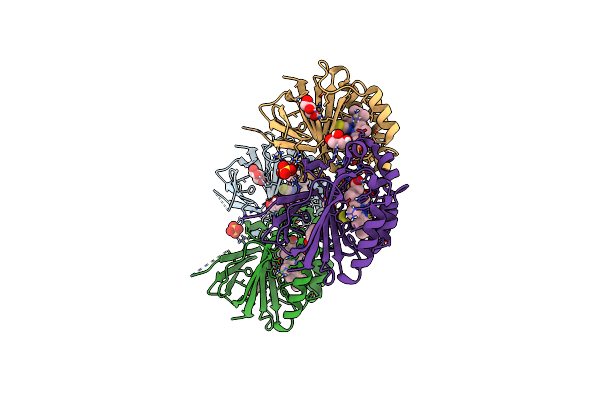
Crystal Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425 (Ph 8.5)
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: MRD, HEM, OH, MPD |
 |
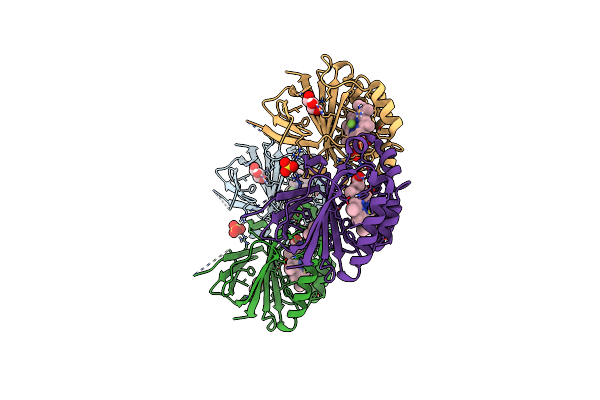
Crystal Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425 In Complex With Isothiocyanate
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: IS8, HEM, GOL, SO4, MG |
 |
Crystal Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425 In Complex With Fluoride
Organism: Cyanothece sp. pcc 7425
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: HEM, F, GOL, SO4 |
 |
Crystal Structure Of Mutant Chlorite Dismutase From Candidatus Nitrospira Defluvii R173E
Organism: Candidatus nitrospira defluvii
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, ACT, EDO |
 |
Crystal Structure Of Mutant Chlorite Dismutase From Candidatus Nitrospira Defluvii W145F In Complex With Cyanide
Organism: Candidatus nitrospira defluvii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, CYN, EDO |
 |
Crystal Structure Of Mutant Chlorite Dismutase From Candidatus Nitrospira Defluvii W145F
Organism: Candidatus nitrospira defluvii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, EDO, GOL |
 |
Crystal Structure Of Mutant Chlorite Dismutase From Candidatus Nitrospira Defluvii W145V
Organism: Candidatus nitrospira defluvii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, IMD, EDO |
 |
Crystal Structure Of Mutant Chlorite Dismutase From Candidatus Nitrospira Defluvii W146Y R173Q
Organism: Candidatus nitrospira defluvii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, EDO, IMD |
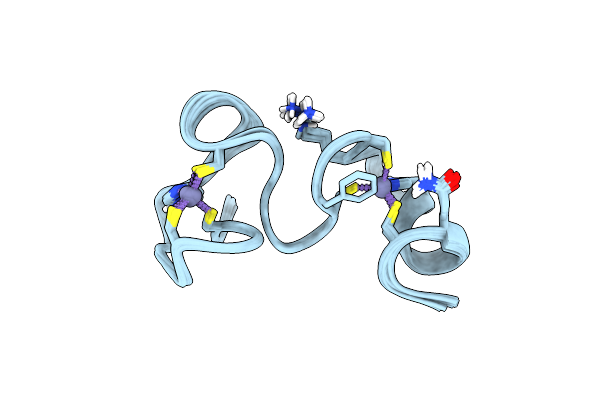 |
Solution Structure Of The Zn Complex Of Eiav Ncp11(22-58) Peptide, Including Two Cchc Zn-Binding Motifs.
Organism: Equine infectious anemia virus
Method: SOLUTION NMR Release Date: 2006-04-10 Classification: NUCLEOCAPSID PROTEIN Ligands: ZN |

