Search Count: 620
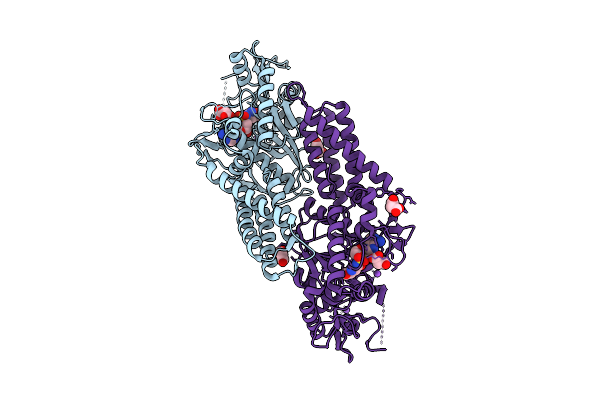 |
Crystal Structure Of Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum In Complex With O5'-(L-Glutamyl-Sulfamoyl)-Adenosine
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: MLI, GSU, ZN, NA |
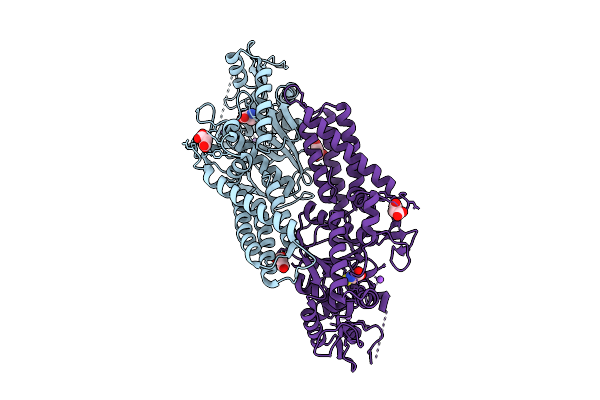 |
Crystal Structure Of Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum In Complex With Cysteine
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: MLI, CYS, ZN, NA |
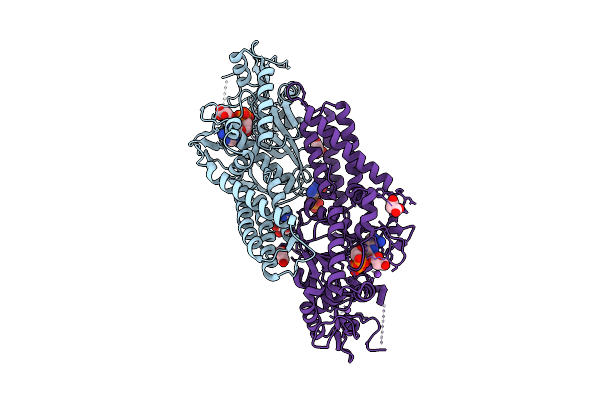 |
Crystal Structure Of Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum In Complex With Adp
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: ADP, MLI, ZN, NA |
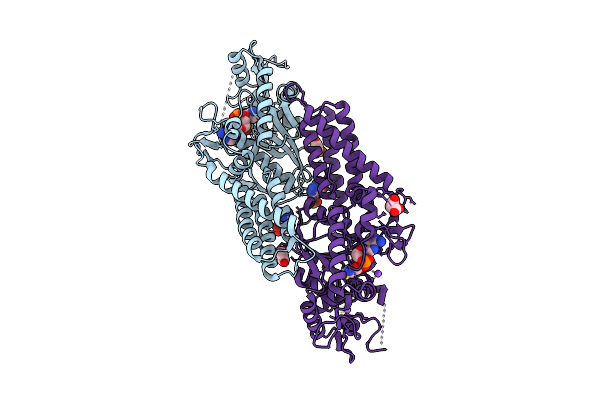 |
Crystal Structure Of Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum In Complex With Amp And Cysteine
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: CYS, AMP, MLI, ZN, NA |
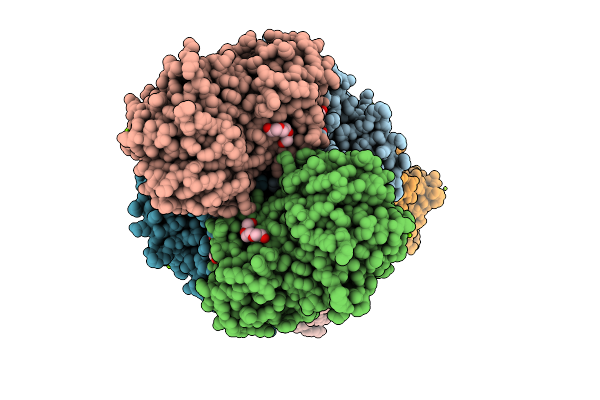 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Bordetella Pertussis (Monoclinic P Form)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: MG, PGE, PG4, PEG, GOL, TRS |
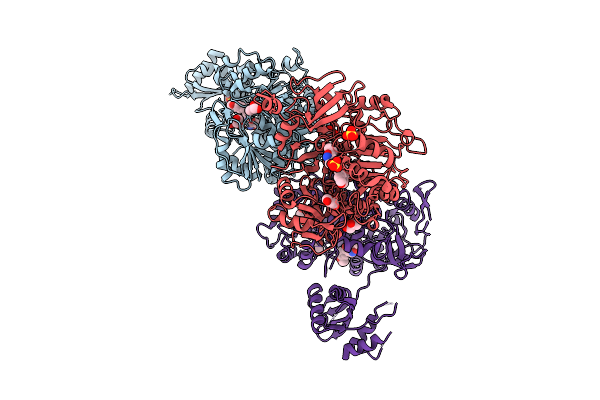 |
Crystal Structure Of Acetyl-Coa Synthetase From Cryptococcus Neoformans H99 In Complex With Inhibitor Hgn-1310 (Dd3-027)
Organism: Cryptococcus neoformans var. grubii
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LIGASE Ligands: GOL, SO4, CL, A1AV1 |
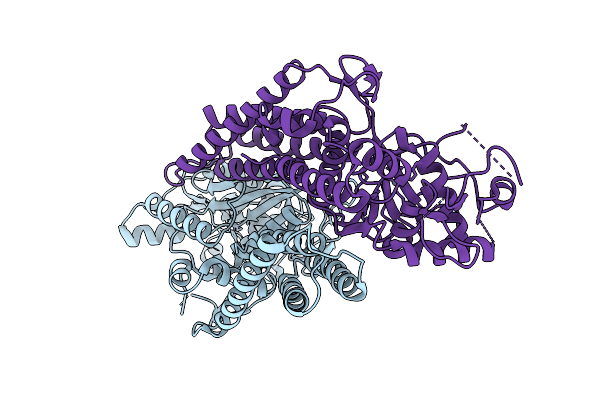 |
Crystal Structure Of Apo Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum (Orthrhombic P Form)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: ZN |
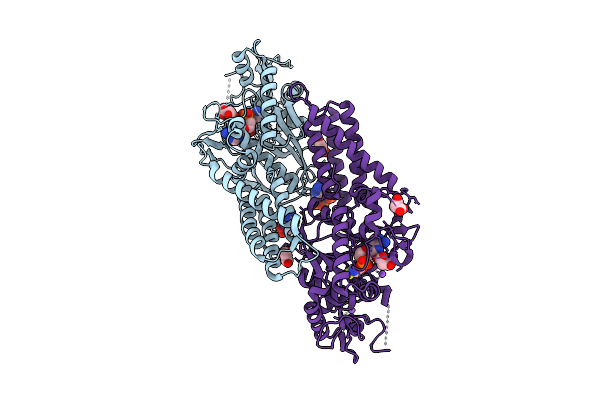 |
Crystal Structure Of Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum In Complex With Cysteinyl-Amp
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: A1CYZ, ATP, MLI, ZN, NA |
 |
Crystal Structure Of Cysteinyl-Trna Synthetase (Cysrs) From Plasmodium Falciparum In Complex With 5'-Sulfamoyladenosine
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: LMS, ZN, NA, CL, MLI |
 |
Crystal Structure Of 6,7-Dimethyl-8-Ribityllumazine Synthase From Bordetella Pertussis In Complex With 5-Amino-6-(D-Ribitylamino)Uracil
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: CL, LMZ |
 |
Crystal Structure Of Glutamate--Trna Ligase (Gltx) From Moraxella Catarrhalis In Complex With 5'-O-(N-Glutamyl)Sulfamoyladeonosine
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: GSU, SO4, P6G, 2PE |
 |
Crystal Structure Of Nucleoside-Diphosphate Kinase Cryptosporidium Parvum (Gmp Complex)
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5GP, MG |
 |
Crystal Structure Of Prolyl-Trna Synthetase (Prors, Proline--Trna Ligase) From Plasmodium Falciparum In Complex With Inhibitor Ynw69
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: CL, SO4, A1CYM |
 |
Organism: Babesia microti strain ri
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: PGE, NA, PG4 |
 |
Crystal Structure Of Glutamate Dehydrogenase From Babesia Microti In Complex With Nadp
Organism: Babesia microti strain ri
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: PG4, NAP |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis In Complex With Utp
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: MG, UTP |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis In Complex With Uridine-5'-Diphosphate-Glucose
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: SO4, MG, UPG, GOL |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis In Complex With Uridine-5'-Diphosphate-Glucose (Twinned Lattice)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: UPG, MG |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis (Sulfate Bound)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: SO4, CL, GOL |
 |
Crystal Structure Of A Phage Catechol 1,2-Dioxygenase Identified From A Soil Metagenomic Survey
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE |

