Search Count: 24
 |
Crystal Structure Of Dna Polymerase Beta With Fapy-Dg Base-Paired With A Dc
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-08-03 Classification: DNA BNDING PROTEIN/DNA |
 |
Crystal Structure Of Dna Polymerase Beta With Fapy-Dg Base-Paired With A Da
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Dna Polymerase Beta With Ring Open Intermediate Fapy-Dg Base-Paired With A Dc
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Dna Polymerase Beta With Ring Open Intermediate Fapy-Dg Base-Paired With A Da
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-08-03 Classification: DNA BINDING/DNA |
 |
Binary Complex Of Dna Polymerase Beta With Fapy-Dg In The Template Position
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Binary Complex Of Dna Polymerase Beta With Ring Open Intermediate Fapy-Dg In The Template Position
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Ternary Complex Of Dna Polymerase Beta With Template Fapy-Dg And An Incoming Dctp Analog
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN/DNA Ligands: XC5, MN, NA, CL |
 |
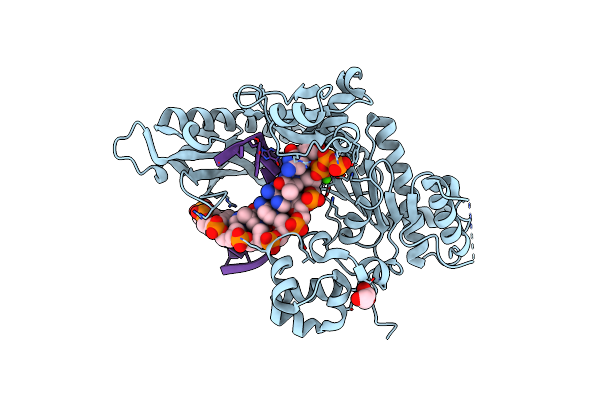
Ternary Complex Of Dna Polymerase Beta With Template Fapy-Dg And An Incoming Datp Analog
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN/DNA |
 |
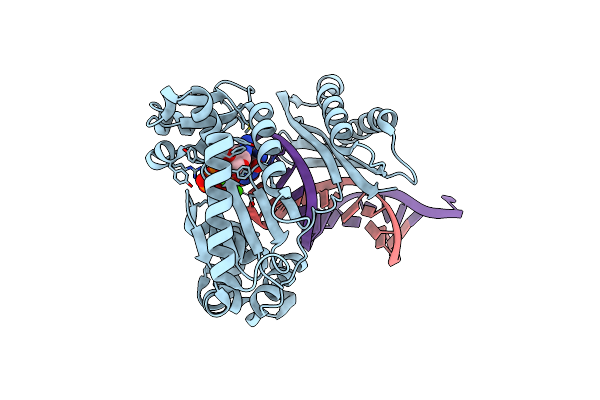
Crystal Structure Of Human Dna Polymerase Eta Inserting Dctp Opposite N-(2'-Deoxyguanosin-8- Yl)-3-Aminobenzanthrone (C8-Dg-Aba)
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-09-07 Classification: transferase/dna Ligands: DCP, CA, GOL |
 |
Base-Displaced Intercalated Structure Of The N-(2'Deoxyguanosin-8-Yl)-3-Aminobenzanthrone Dna Adduct
|
 |
Organism: Sulfolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-09-16 Classification: TRANSFERASE/DNA Ligands: CA, NA, TRS, DCP, GOL |
 |
Dpo4 Post-Insertion Complex With The N-(Deoxyguanosin-8-Yl)-1-Aminopyrene Lesion
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-06-12 Classification: TRANSFERASE/DNA Ligands: DGT, CA, GTP |
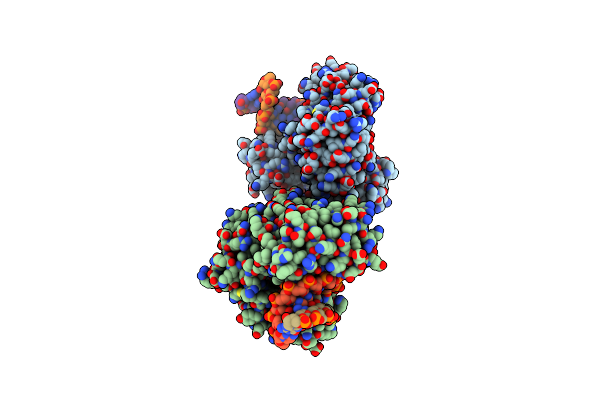 |
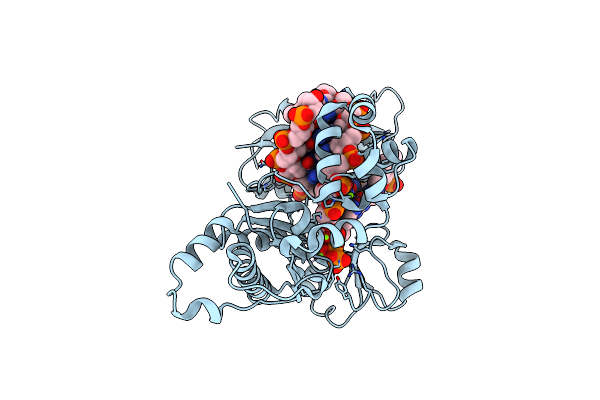
Dpo4 Polymerase Pre-Insertion Binary Complex With The N-(Deoxyguanosin-8-Yl)-1-Aminopyrene Lesion
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-06-12 Classification: TRANSFERASE/DNA Ligands: CA |
 |
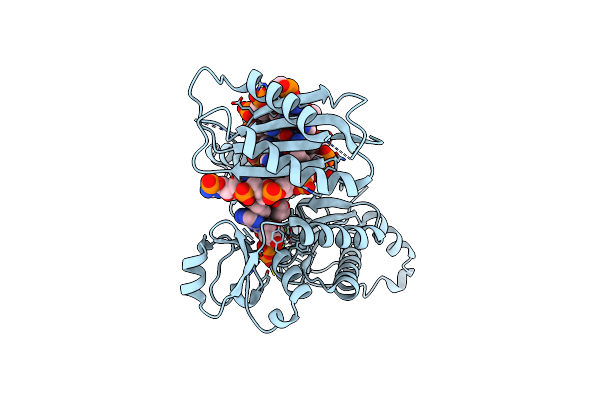
Human Dna Polymerase Iota Incorporating Dctp Opposite N-(Deoxyguanosin-8-Yl)-1-Aminopyrene Lesion
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-01-09 Classification: TRANSFERASE/DNA Ligands: DCP, MG |
 |
Human Dna Polymerase Iota Incorporating Datp Opposite N-(Deoxyguanosin-8-Yl)-1-Aminopyrene Lesion
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-01-09 Classification: TRANSFERASE/DNA Ligands: DTP, MG |
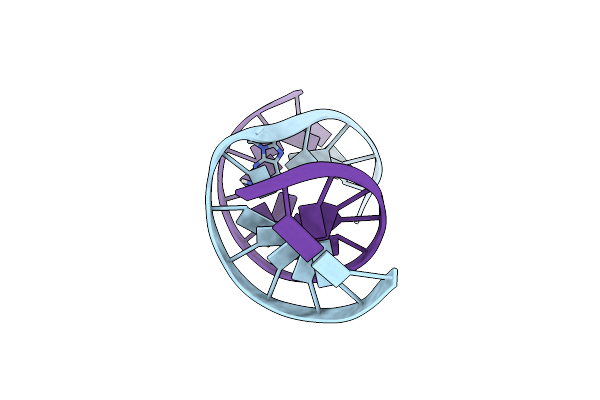 |
Structure Of The Duplex When (5'S)-8,5'-Cyclo-2'-Deoxyguanosine Is Placed Opposite Dt
|
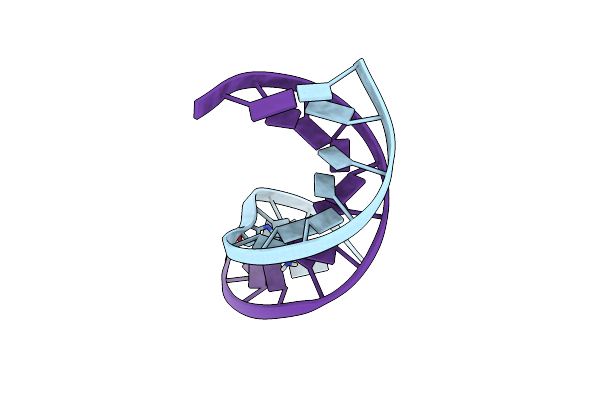 |
Structure Of The Duplex When (5'S)-8,5'-Cyclo-2'-Deoxyguanosine Is Placed Opposite Da
|
 |
 |
 |
Solution Structure Of Cis-5R,6S-Thymine Glycol Opposite Complementary Adenine In Duplex Dna
|

