Search Count: 14
 |
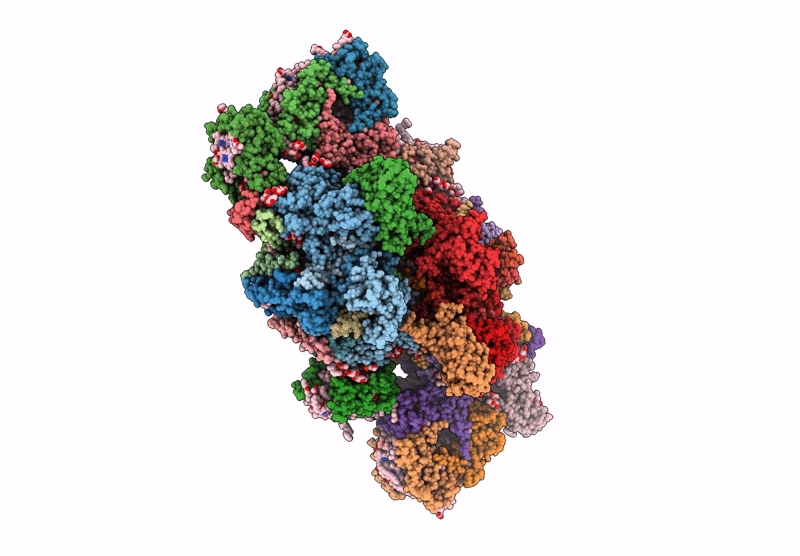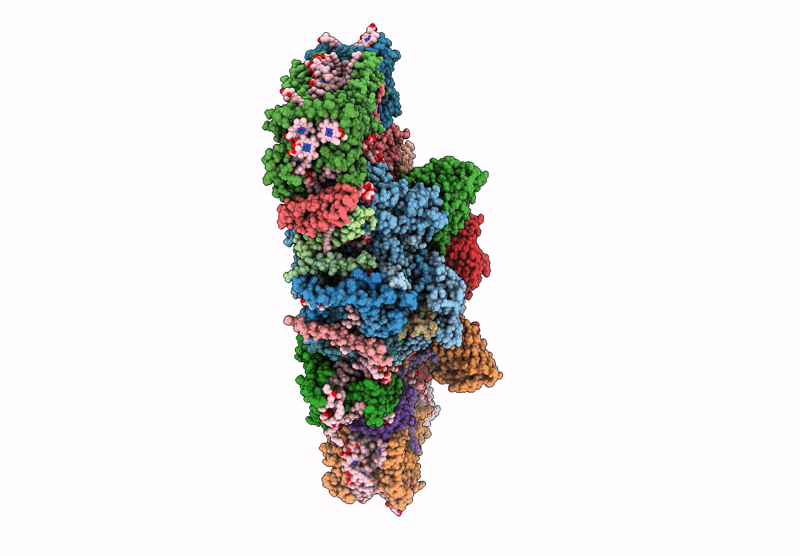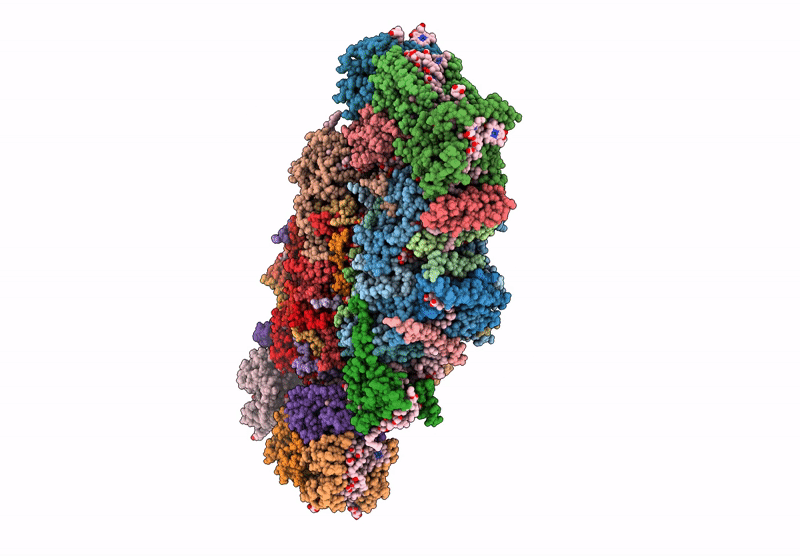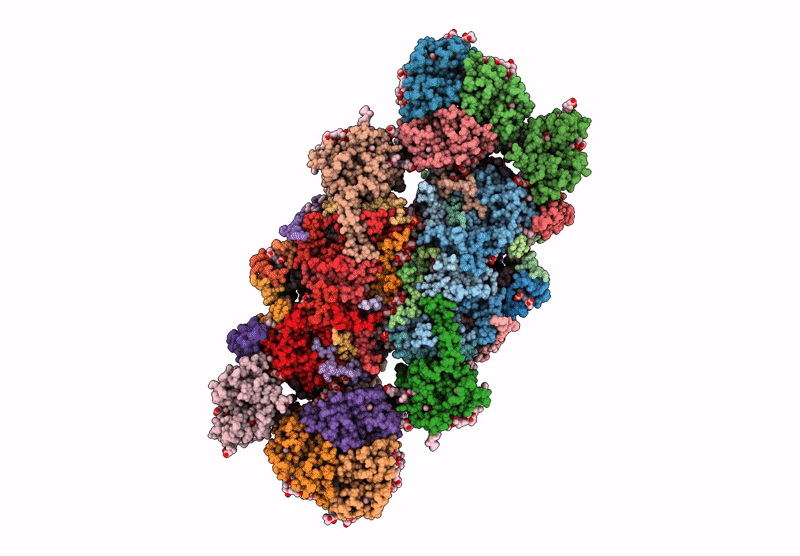
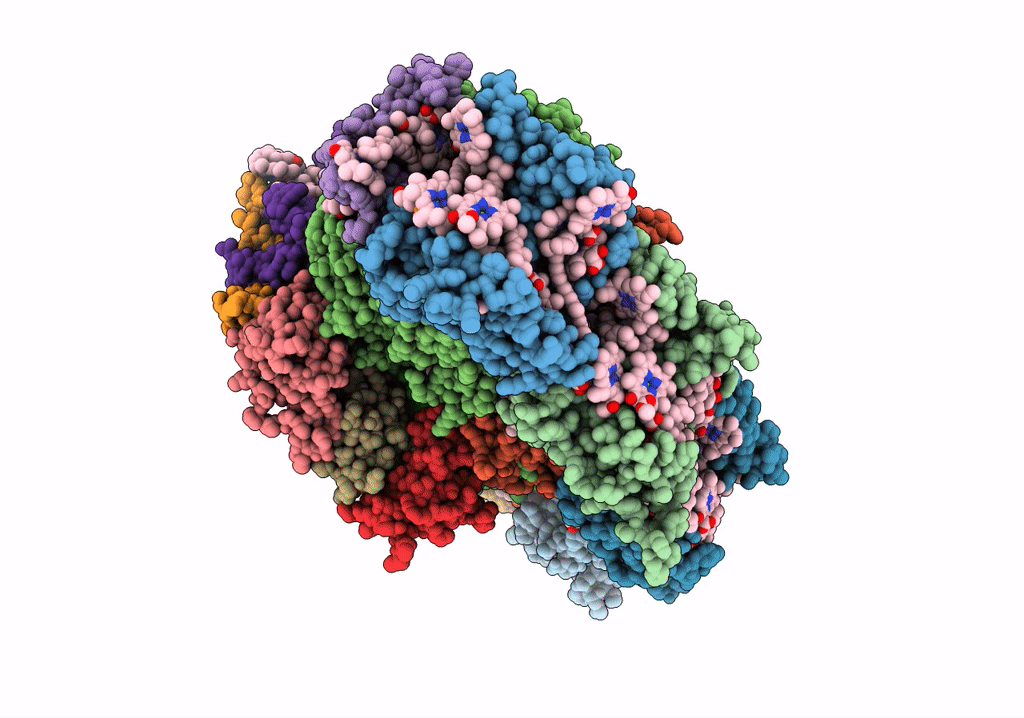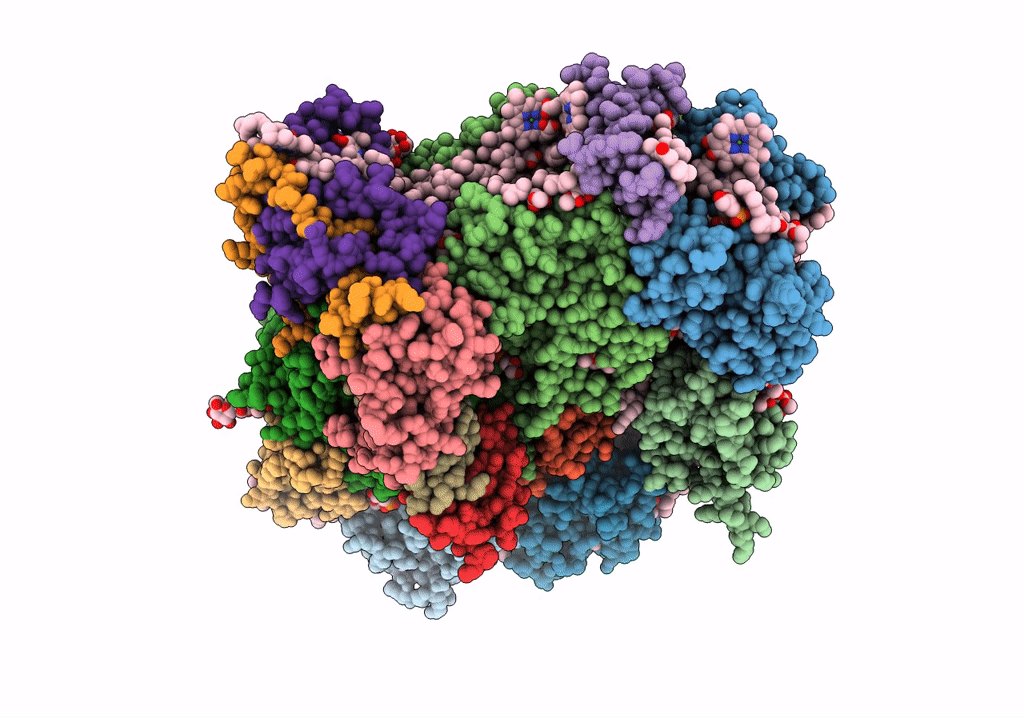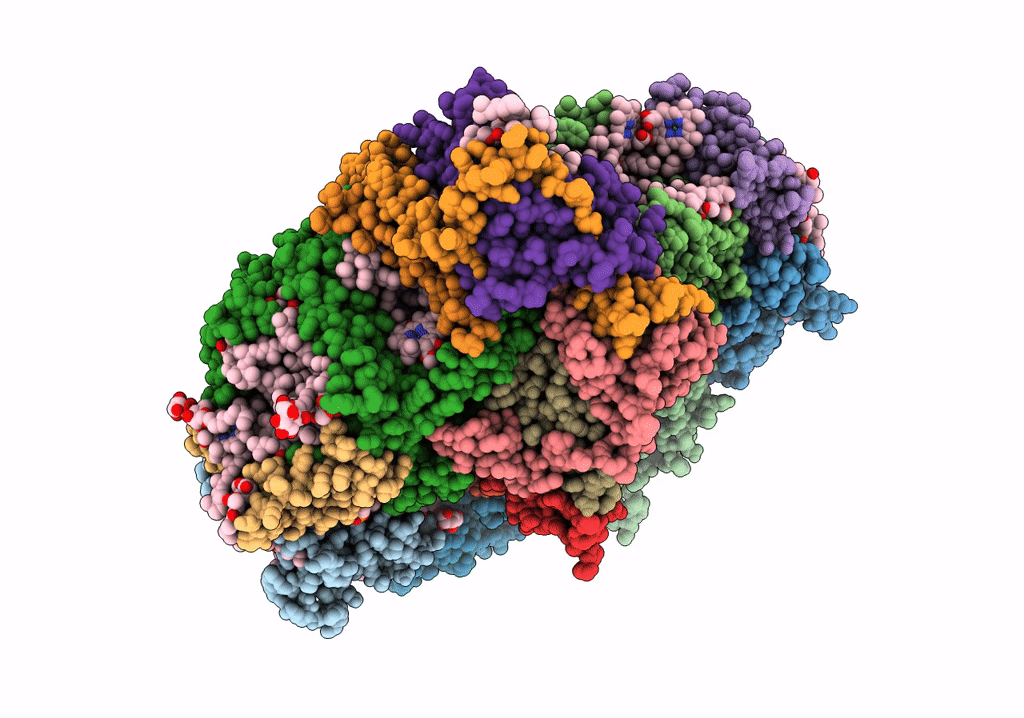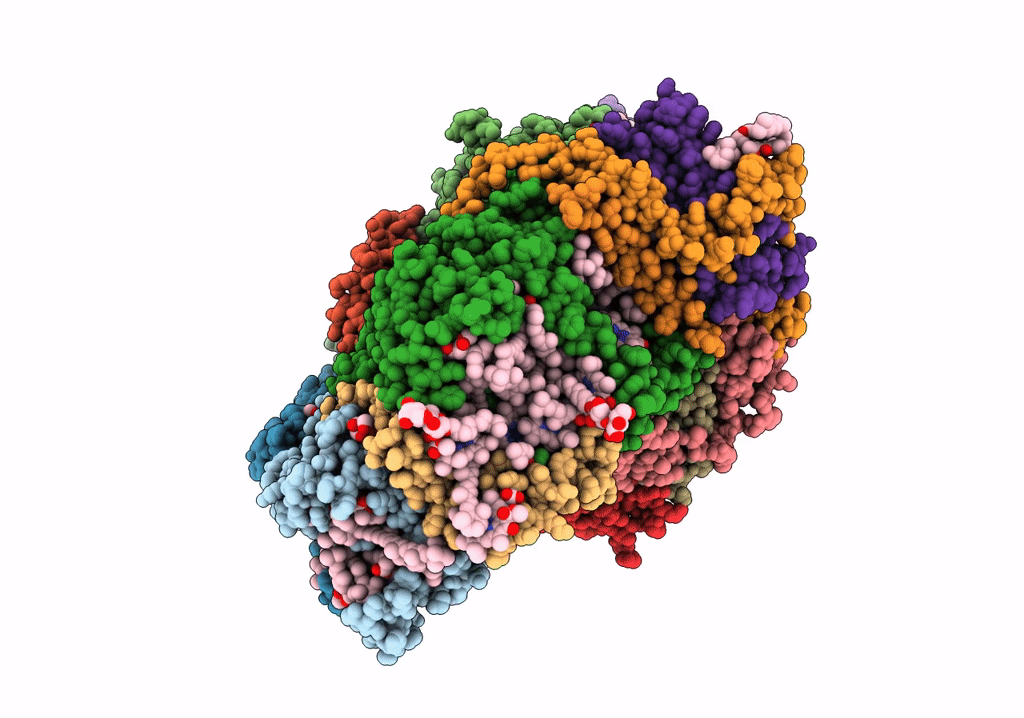
Cryo-Em Structure Of Lhcb4.1-C2S2 Psii-Lhcii Supercomplex From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: PHOTOSYNTHESIS Ligands: CLA, PHO, BCR, SQD, LMG, DGD, FE2, LHG, BCT, PL9, HEM, CHL, LUT, NEX, XAT |
 |
The Structure Of Lhcb8-C2S2 Psii-Lhcii Supercomplex From Arabidopsis Thaliana
|
 |
Left Psi In The Cyclic Electron Transport Supercomplex Ndh-Psi From Arabidopsis
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: ELECTRON TRANSPORT Ligands: CLA, PQN, LHG, BCR, SF4, LMU, DGD, LUT, LMG, CHL, XAT |
 |
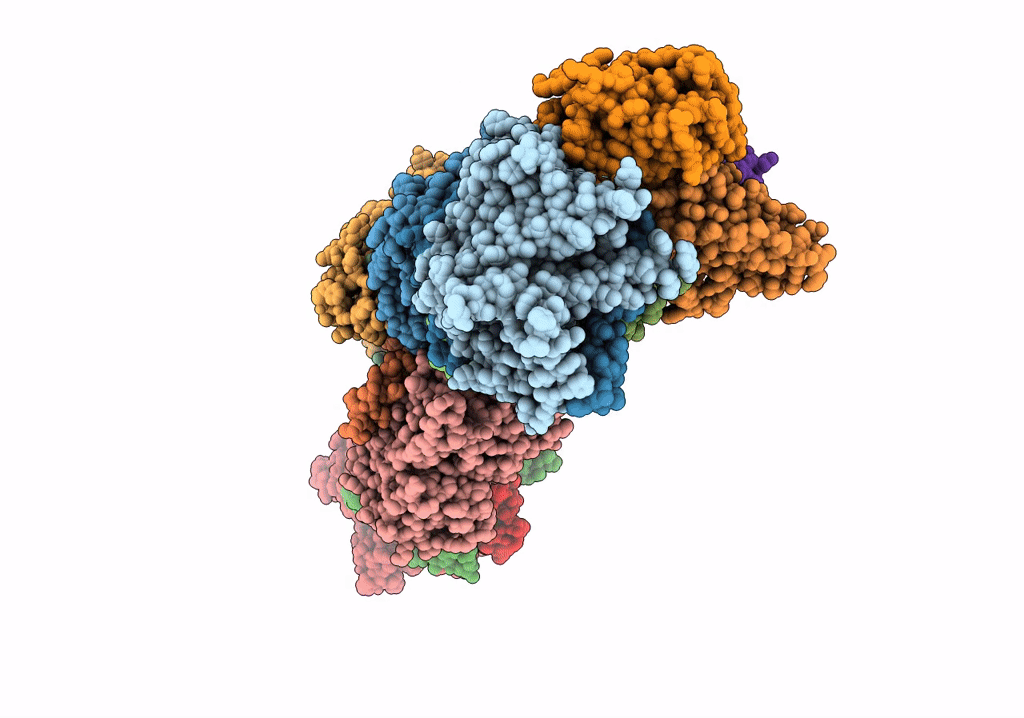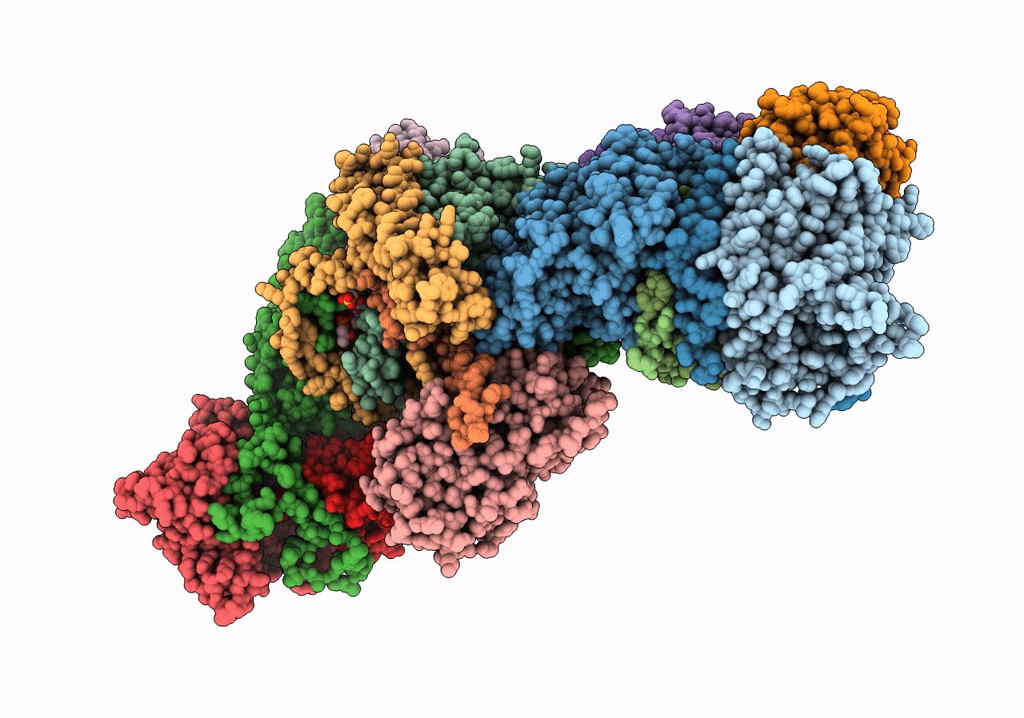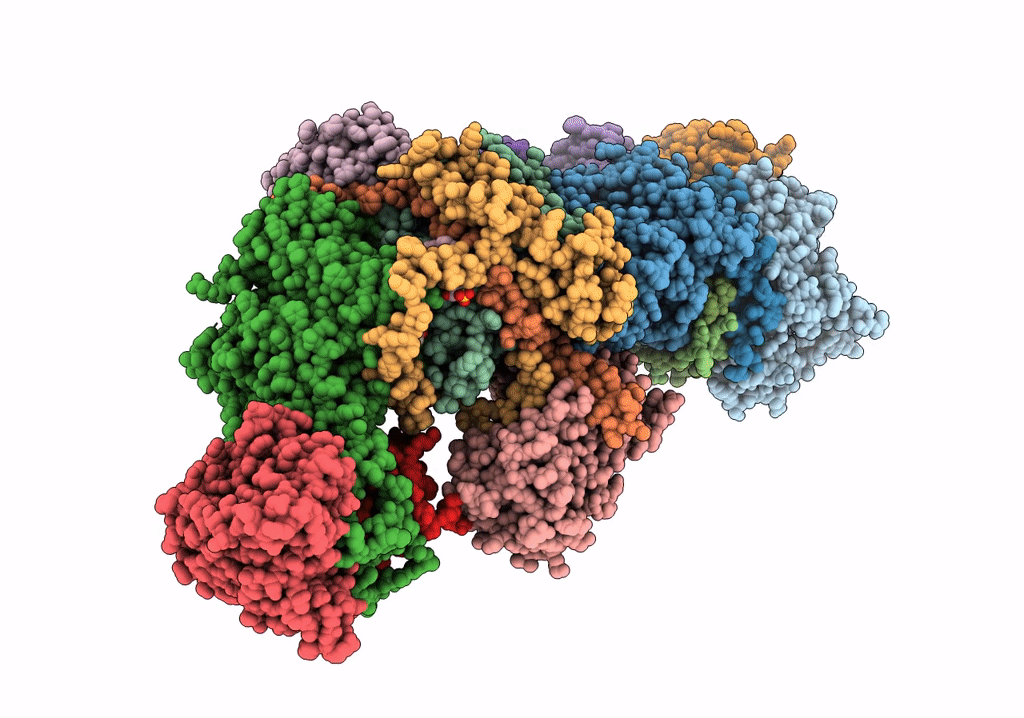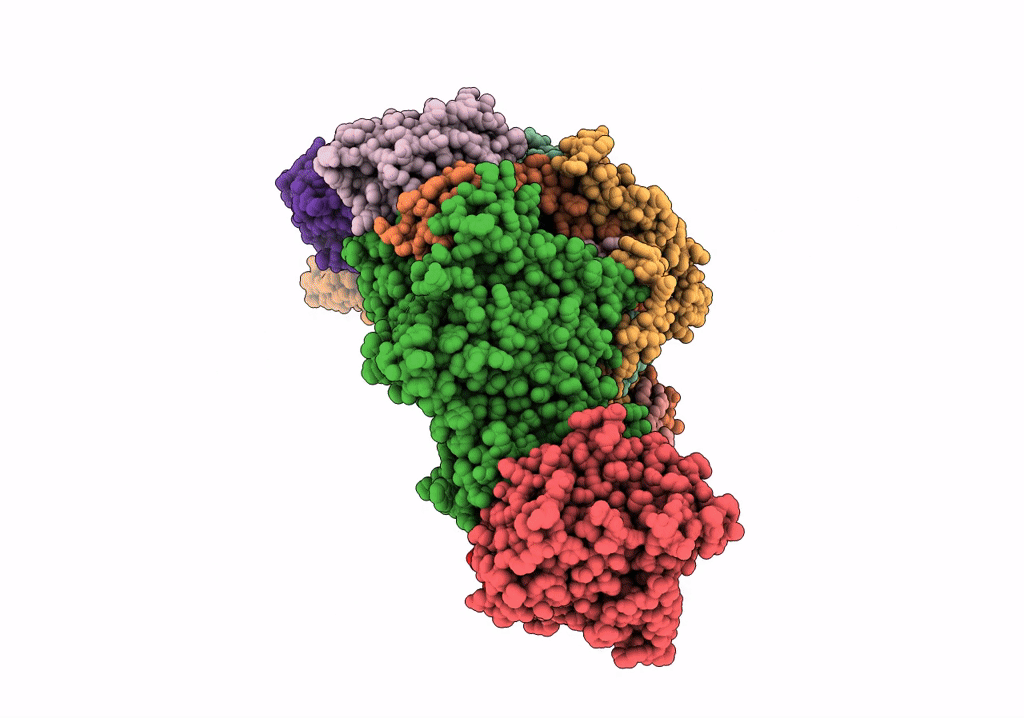
Right Psi In The Cyclic Electron Transfer Supercomplex Ndh-Psi From Arabidopsis
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: ELECTRON TRANSPORT Ligands: CLA, PQN, LHG, BCR, SF4, LMU, DGD, SQD, CHL, LUT, XAT, LMG |
 |
Subcomplexes B,M And L In The Cylic Electron Transfer Supercomplex Ndh-Psi From Arabidopsis
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: ELECTRON TRANSPORT Ligands: LHG, SQD, FES |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: ELECTRON TRANSPORT Ligands: CLA, PQN, LHG, BCR, SF4, LMU, DGD, LUT, LMG, CHL, XAT, SQD, FES |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-05-30 Classification: TRANSFERASE Ligands: NI, AGS, MG |
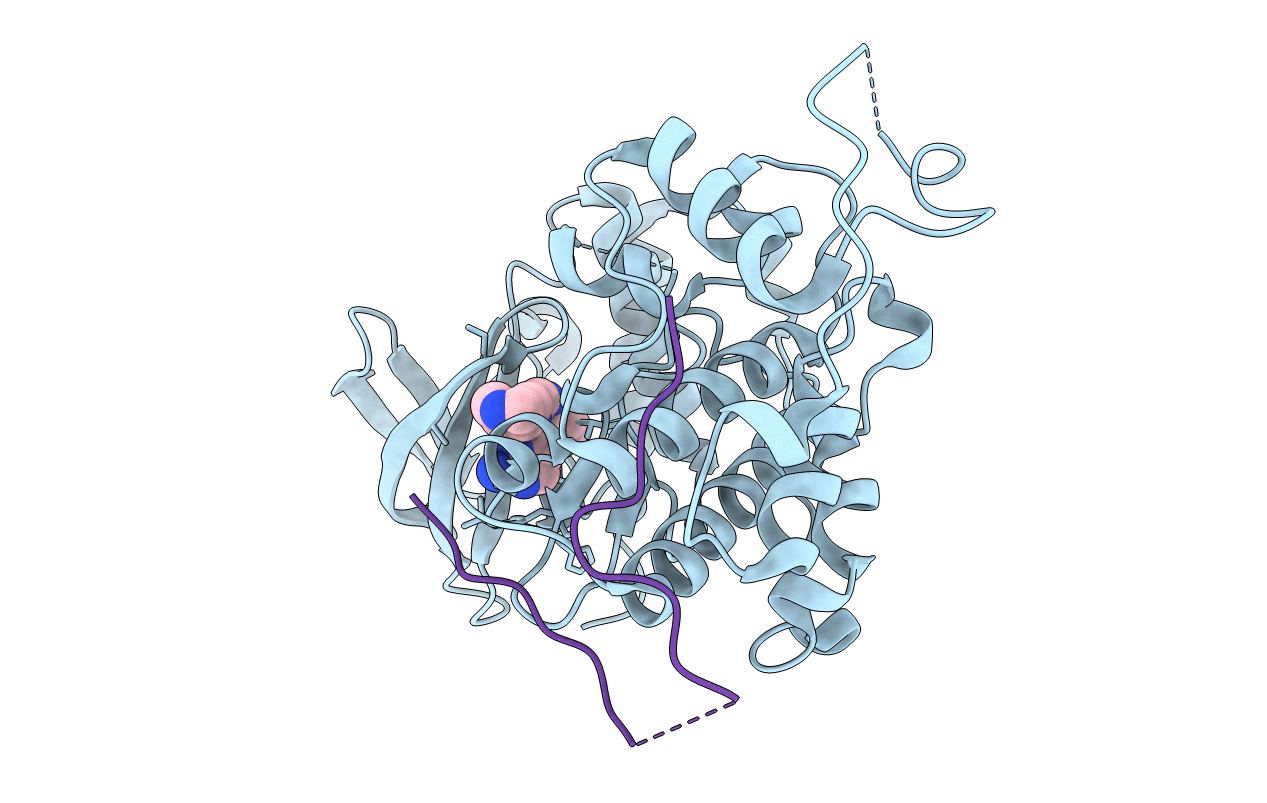 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: SB4 |
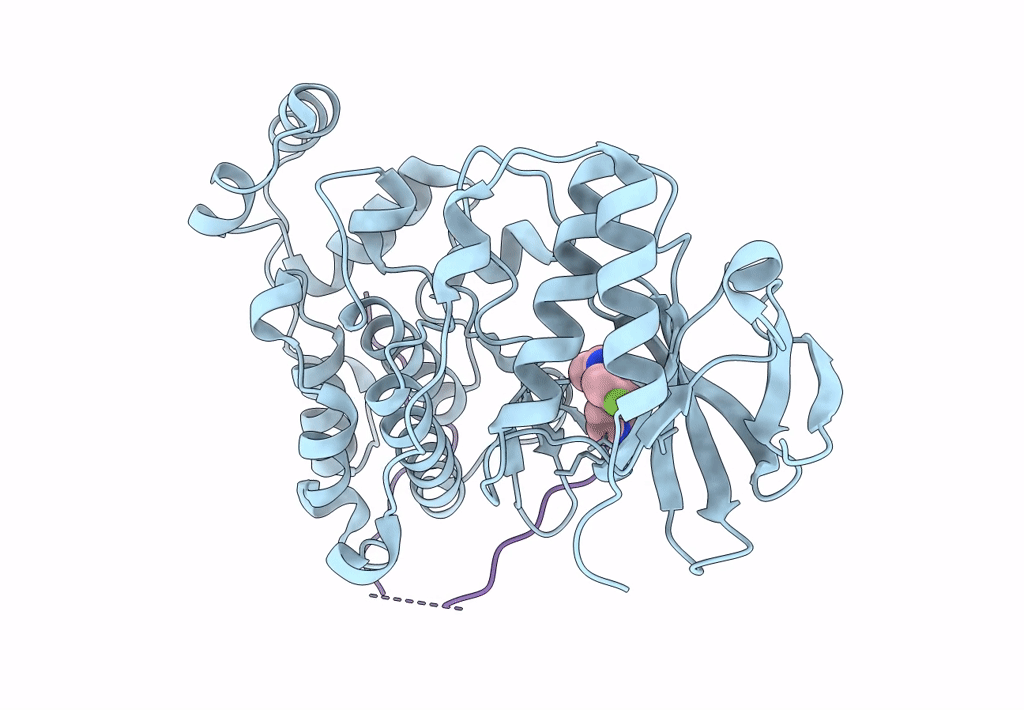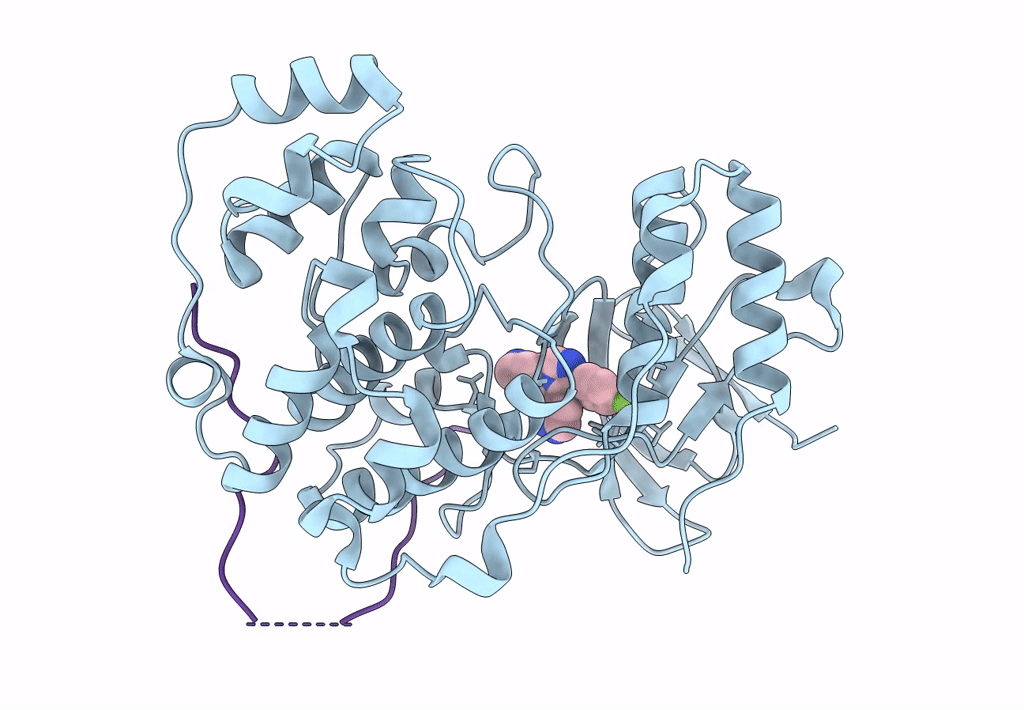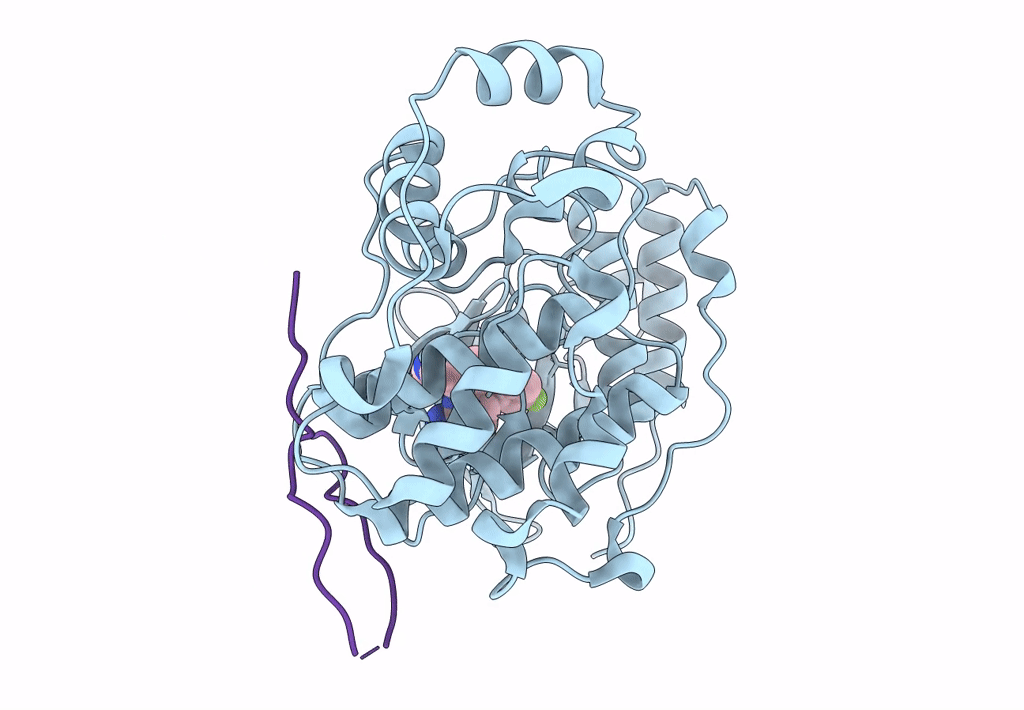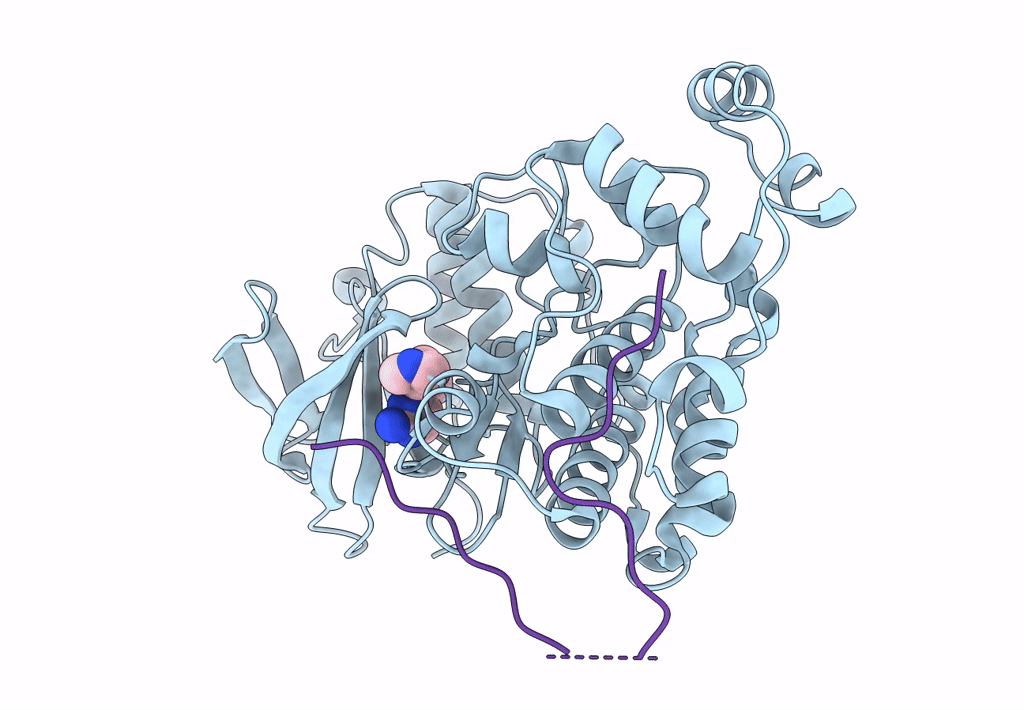 |
Structural Basis Of Autoactivation Of P38 Alpha Induced By Tab1 (Monoclinic Crystal Form)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-08-21 Classification: TRANSFERASE Ligands: SB4 |
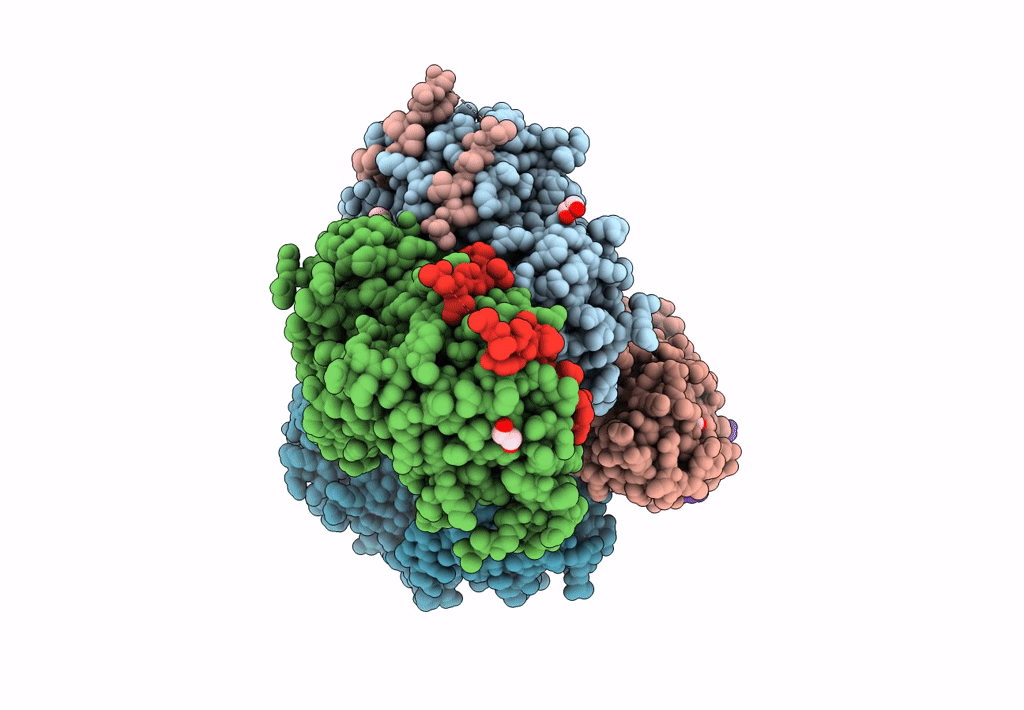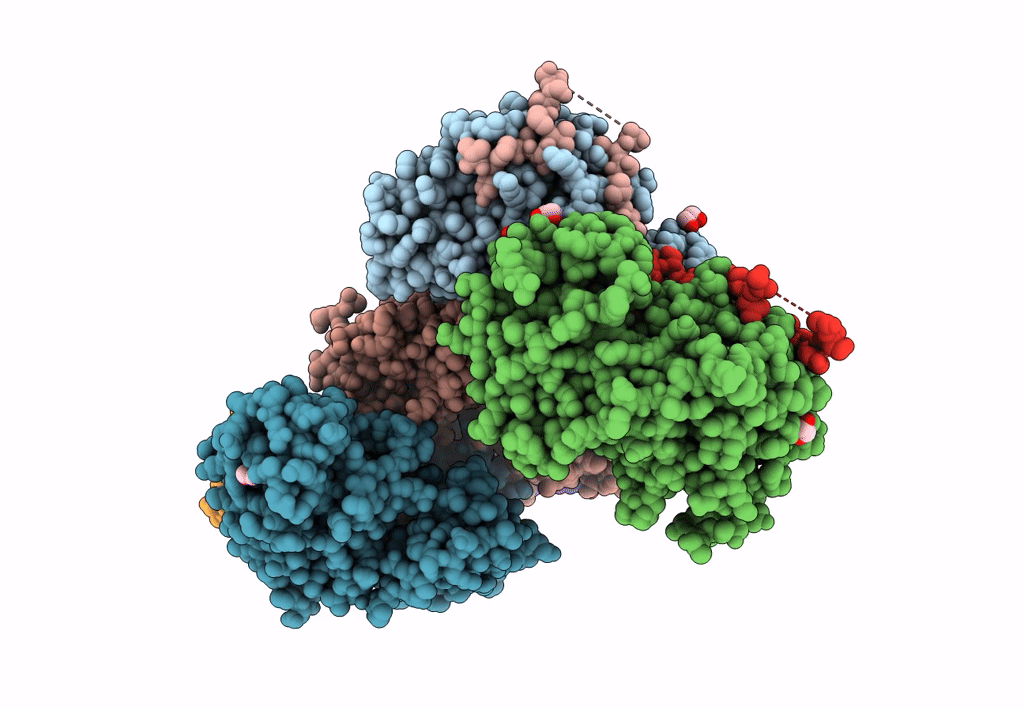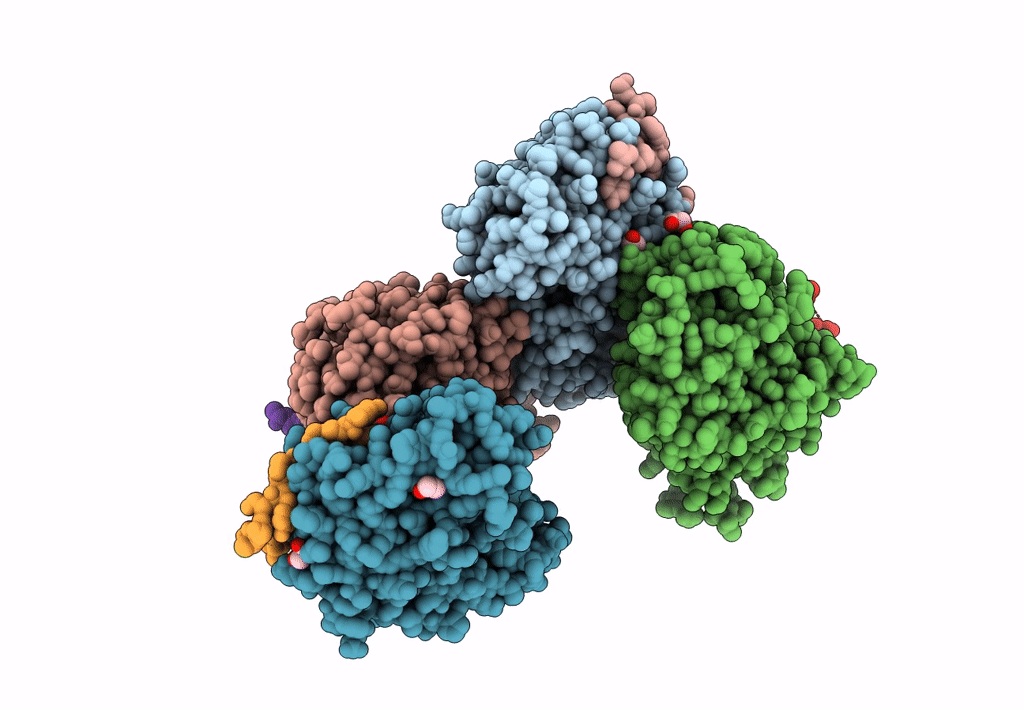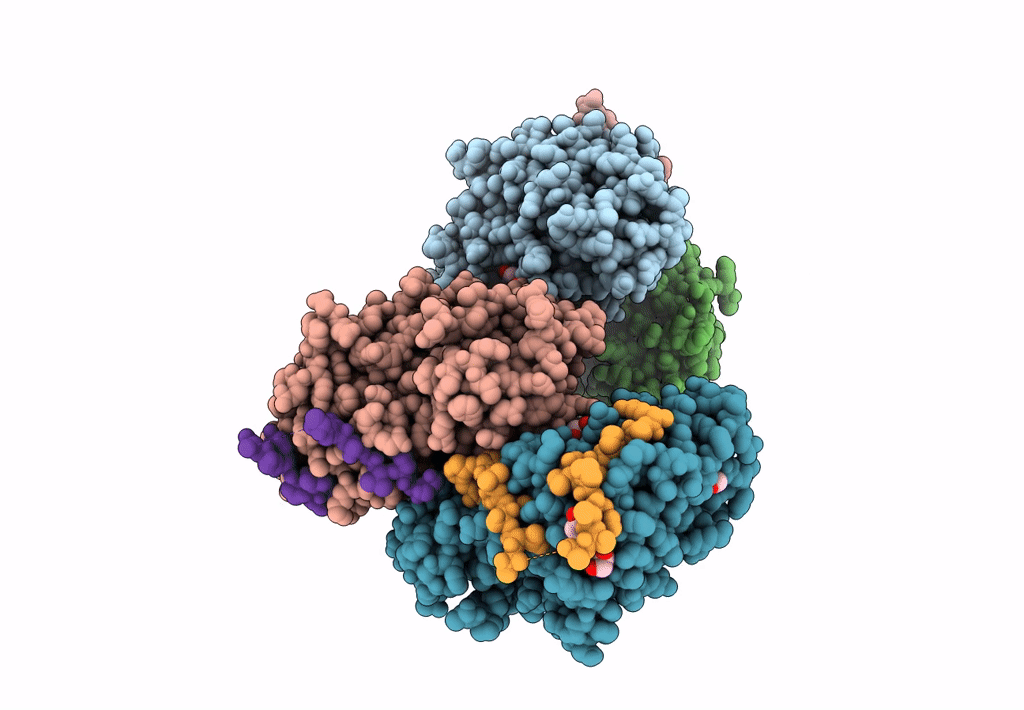 |
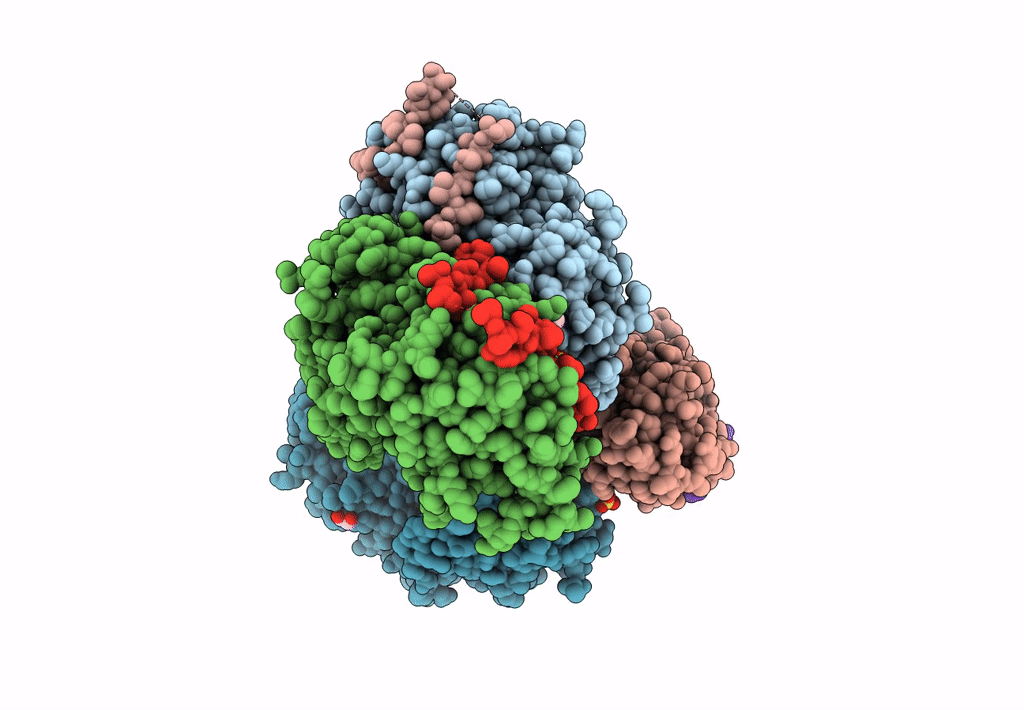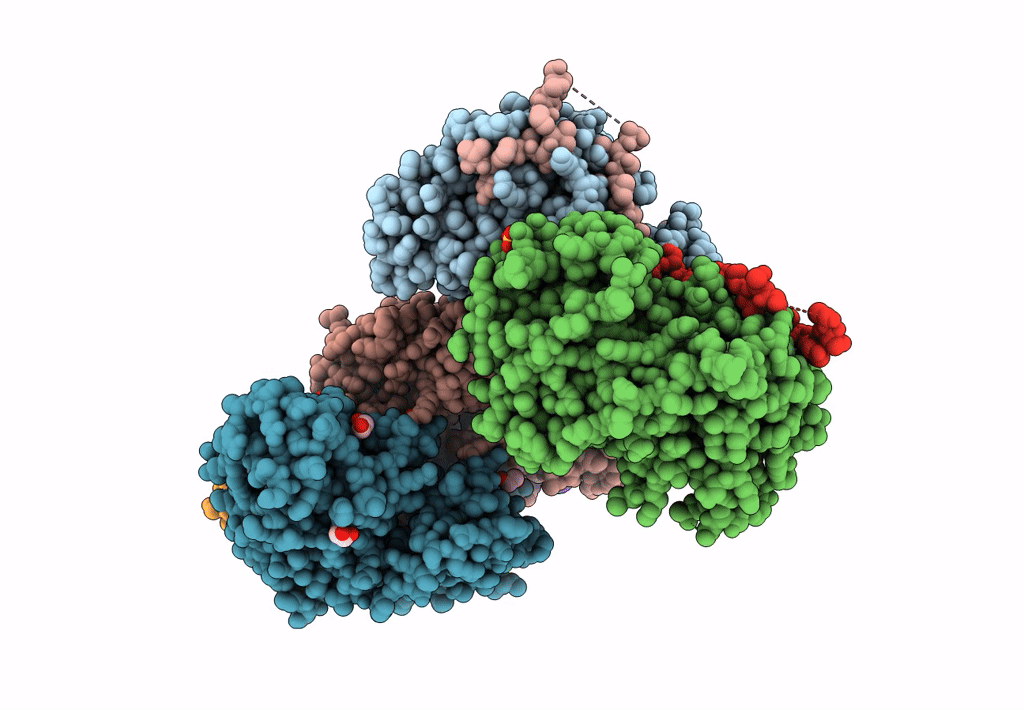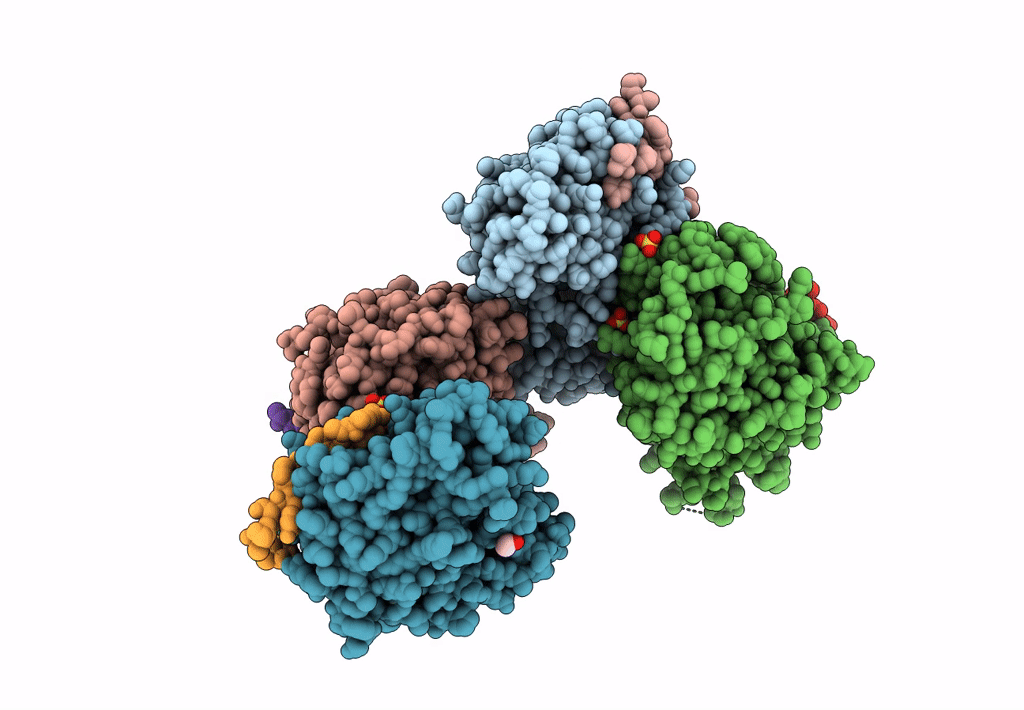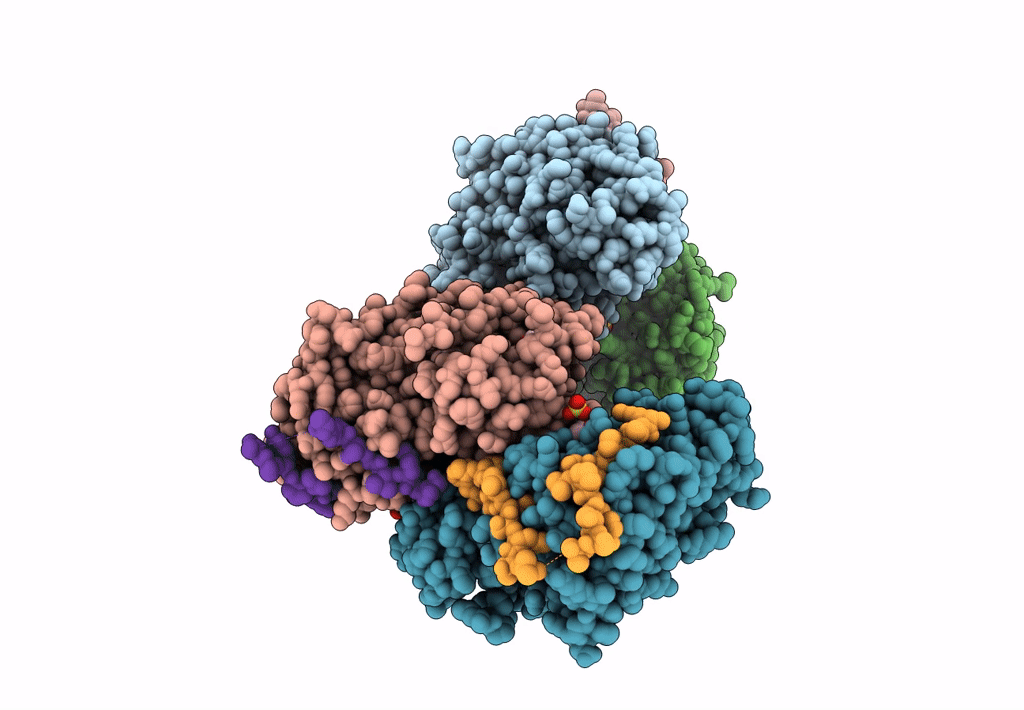
Structural Basis Of Autoactivation Of P38 Alpha Induced By Tab1 (Tetragonal Crystal Form)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-08-21 Classification: TRANSFERASE Ligands: SB4, TLA, EDO |
 |
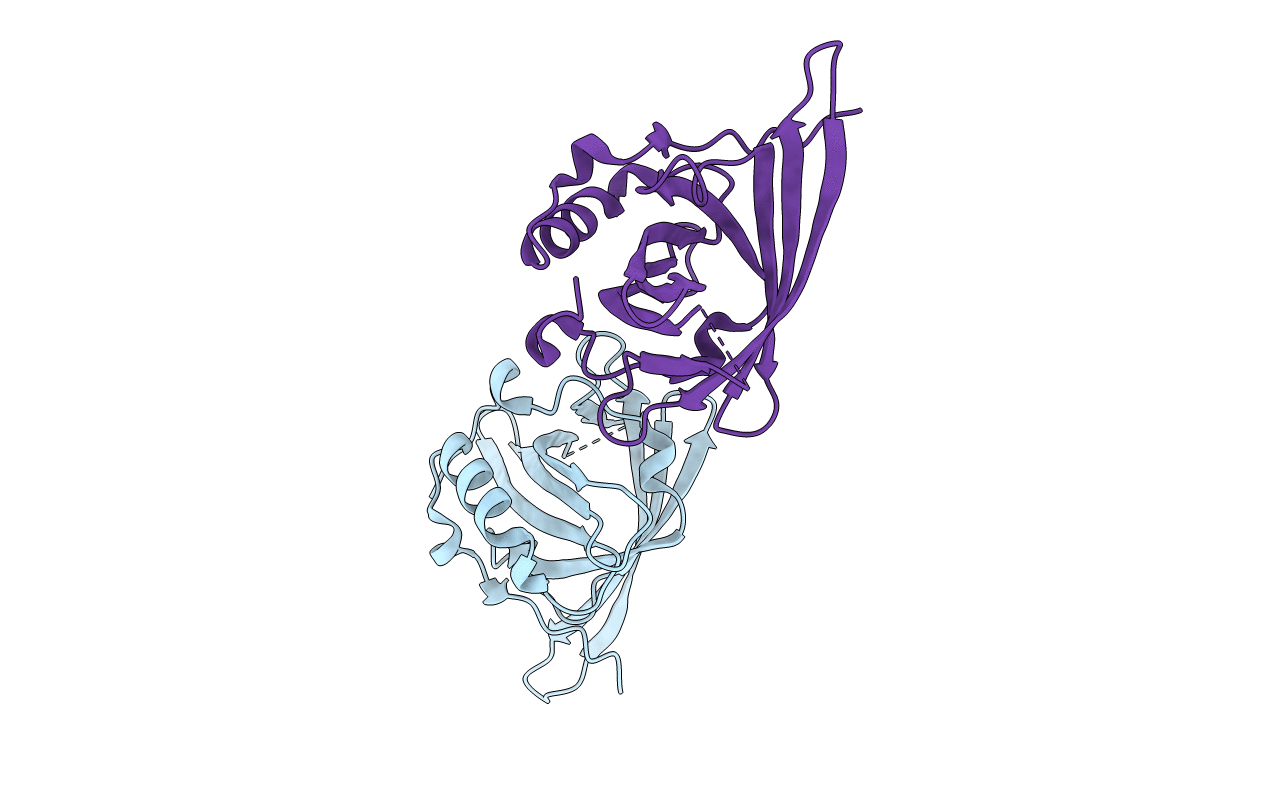
Structural Basis Of Autoactivation Of P38 Alpha Induced By Tab1 (Tetragonal Crystal Form With Bound Sulphate)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2013-08-21 Classification: TRANSFERASE Ligands: SB4, SO4, EDO |
 |
Crystal Structure Of The Lipocalin Domain Of Violaxanthin De-Epoxidase (Vde) At Ph7
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-21 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The Lipocalin Domain Of Violaxanthin De-Epoxidase (Vde) At Ph5
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-21 Classification: OXIDOREDUCTASE Ligands: GD |

