Search Count: 8
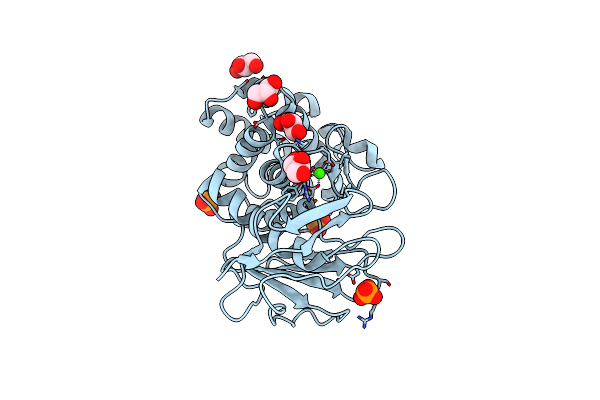 |
Crystal Structure Of Pseudomonas Aeruginosa Strain K Solvent Tolerant Elastase
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: GOL, PO4, CA, ZN |
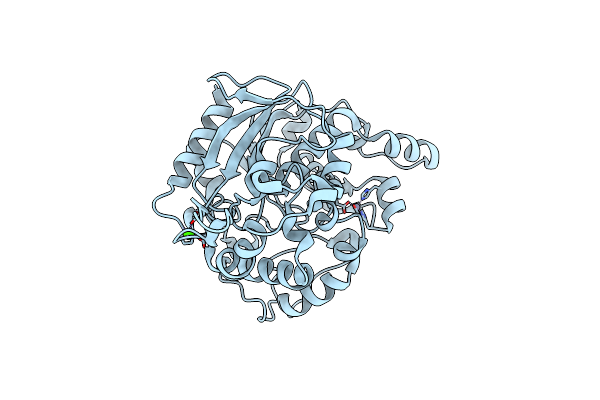 |
Crystallization And 3D Structure Elucidation Of Thermostable L2 Lipase From Thermophilic Locally Isolated Bacillus Sp. L2.
Organism: Bacillus sp. l2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: ZN, CA |
 |
 |
 |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: ZN, GOL, CL, NA, CA |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-10-30 Classification: HYDROLASE Ligands: ZN, CA, CL |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-07-17 Classification: HYDROLASE Ligands: ZN, CA, NA, CL |

