Search Count: 27
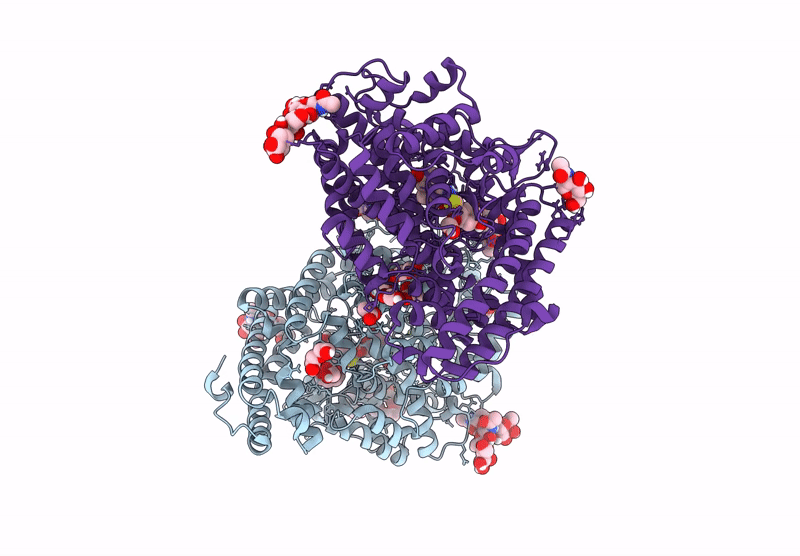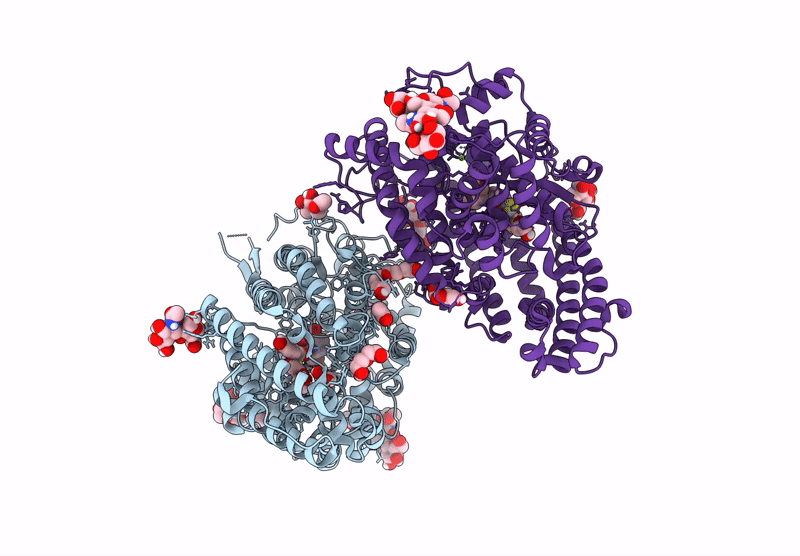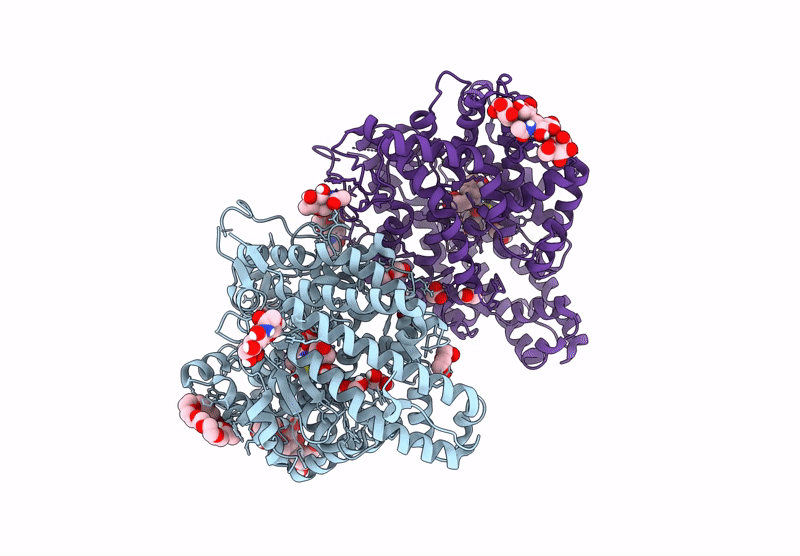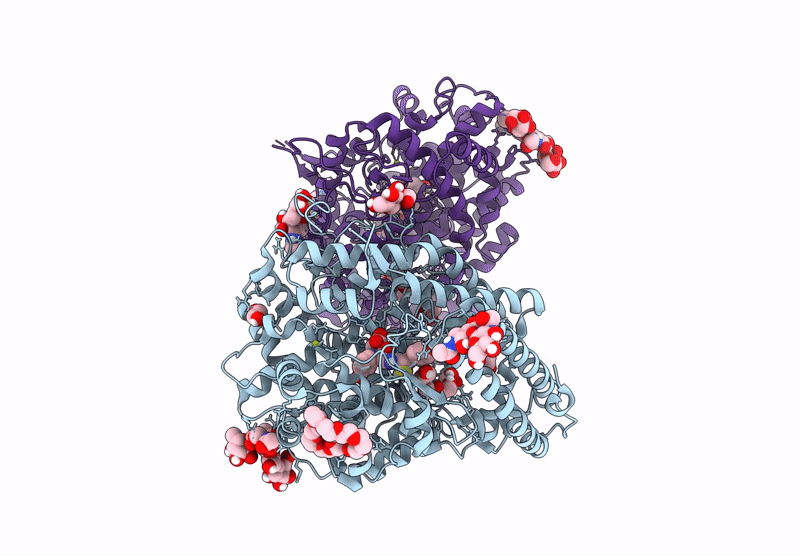 |
Crystal Structure Of Angiotensin-1 Converting Enzyme N-Domain In Complex With Dual Ace/Nep Inhibitor Ad014
|
 |
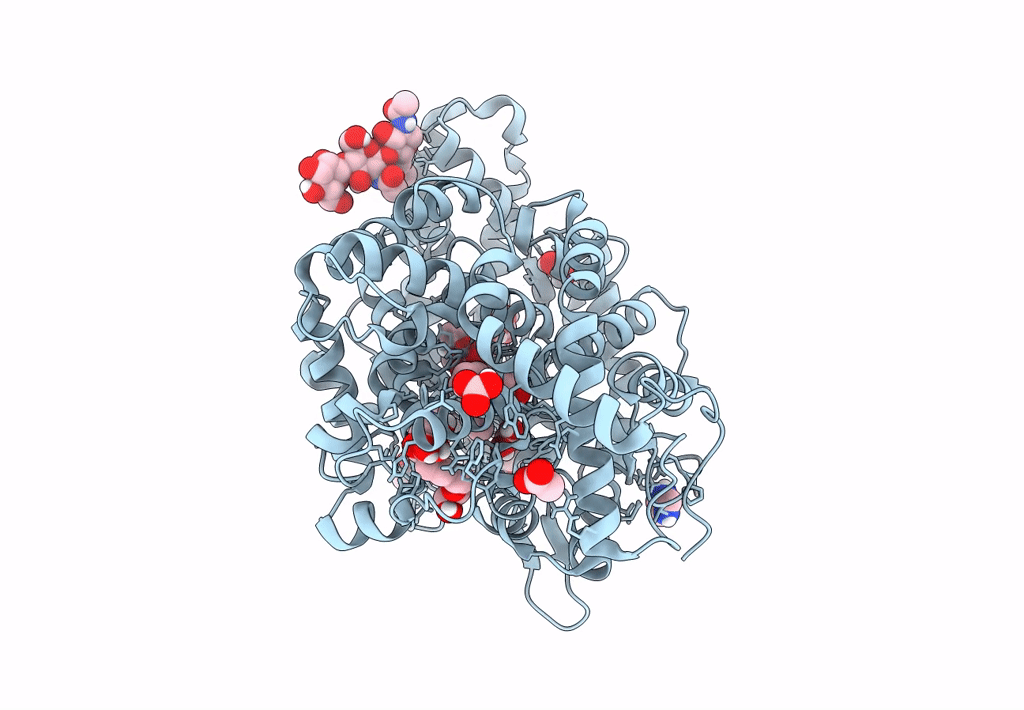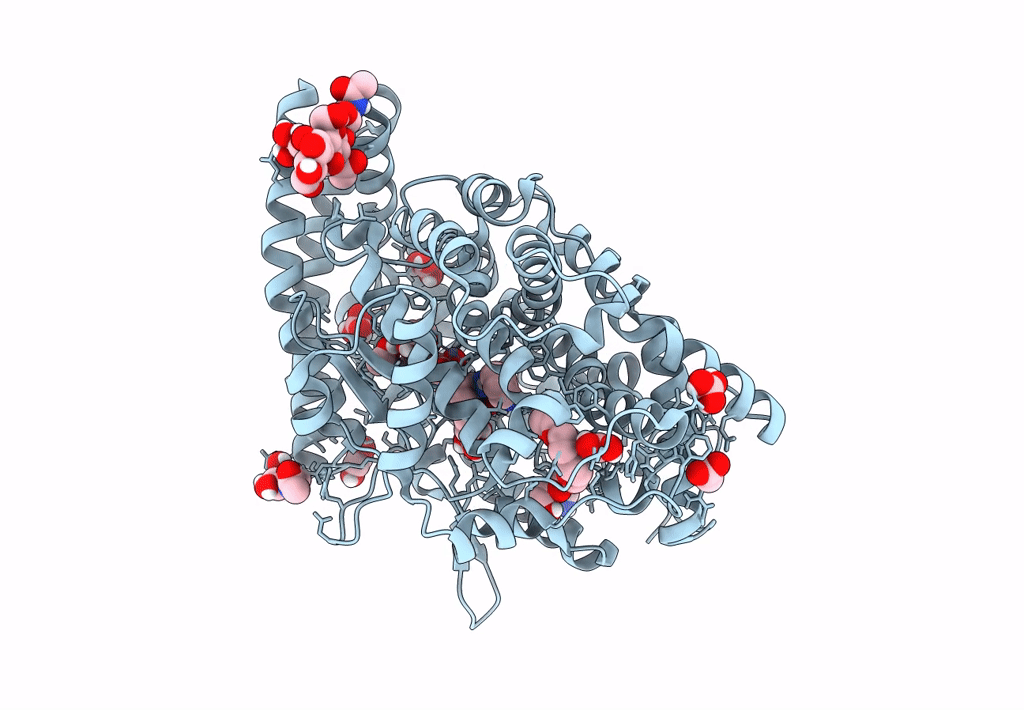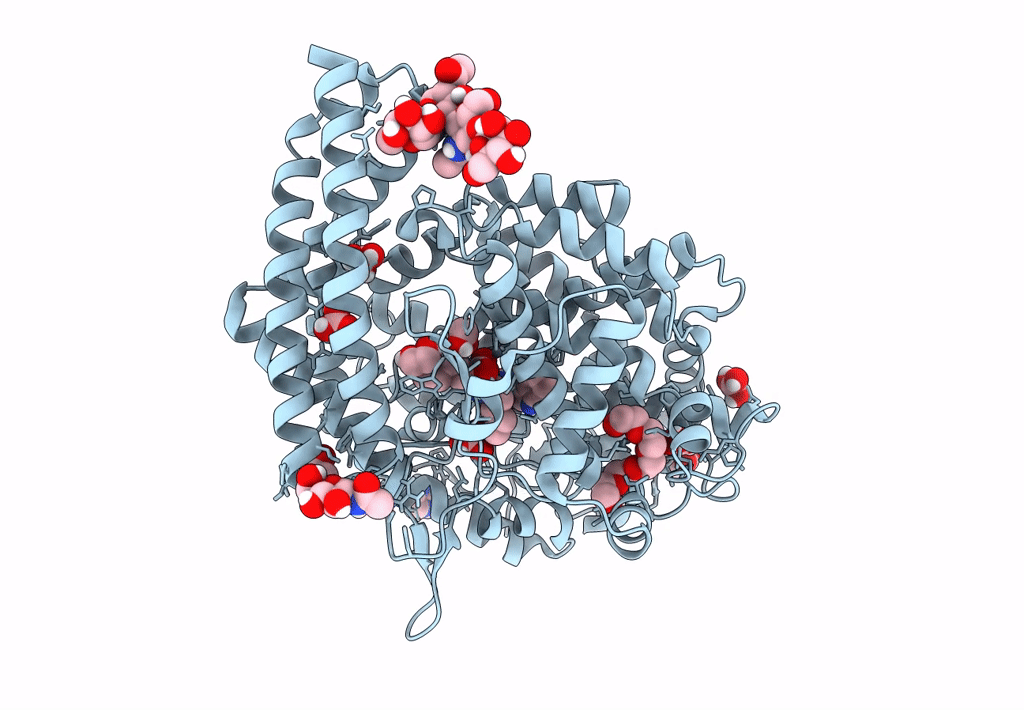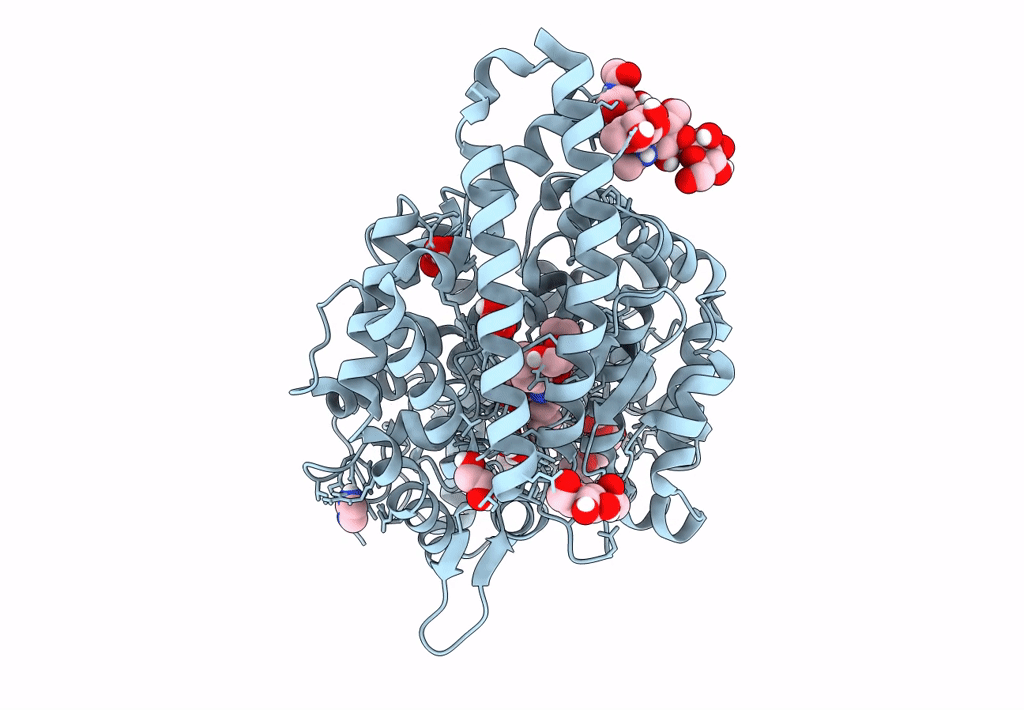
Crystal Structure Of Angiotensin-1 Converting Enzyme N-Domain In Complex With Dual Ace/Nep Inhibitor Ad015
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-04-16 Classification: HYDROLASE Ligands: NAG, PG4, ACY, EDO, PGE, ZN, CL, A1IRR, MG, PEG, PE8 |
 |
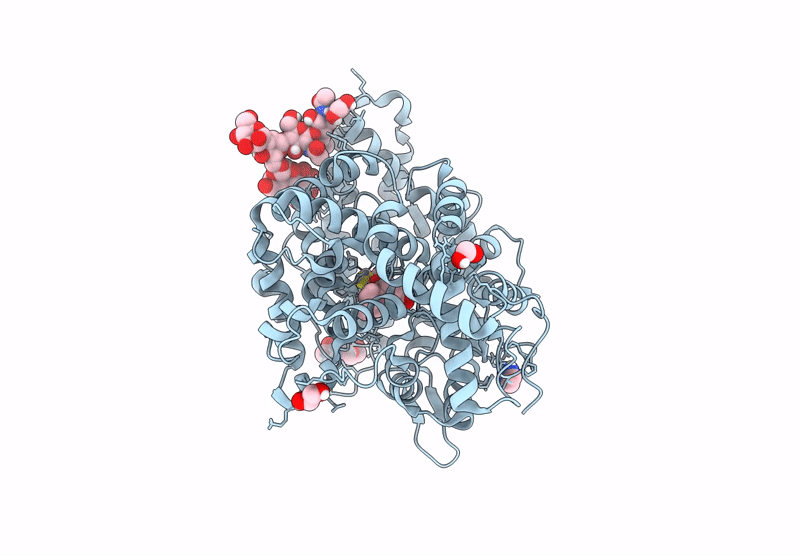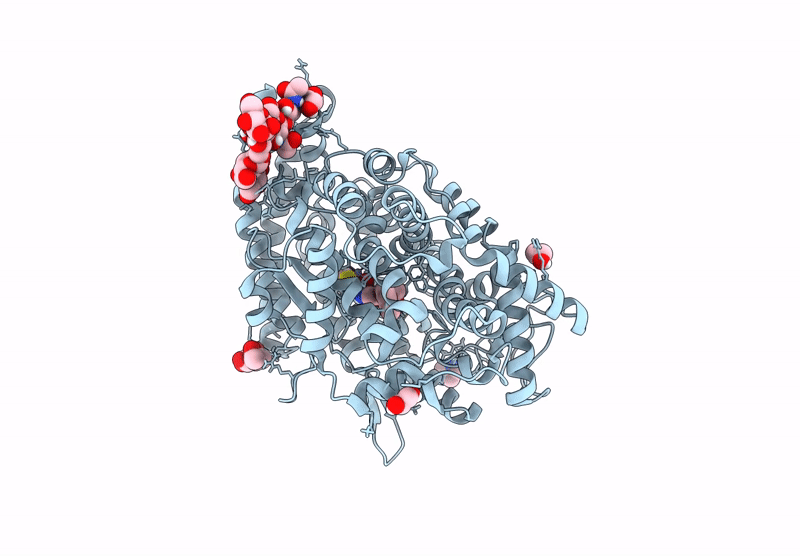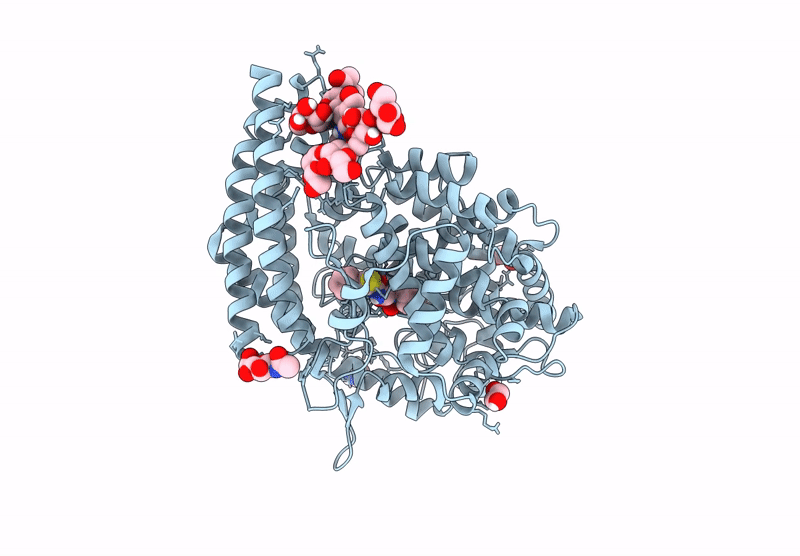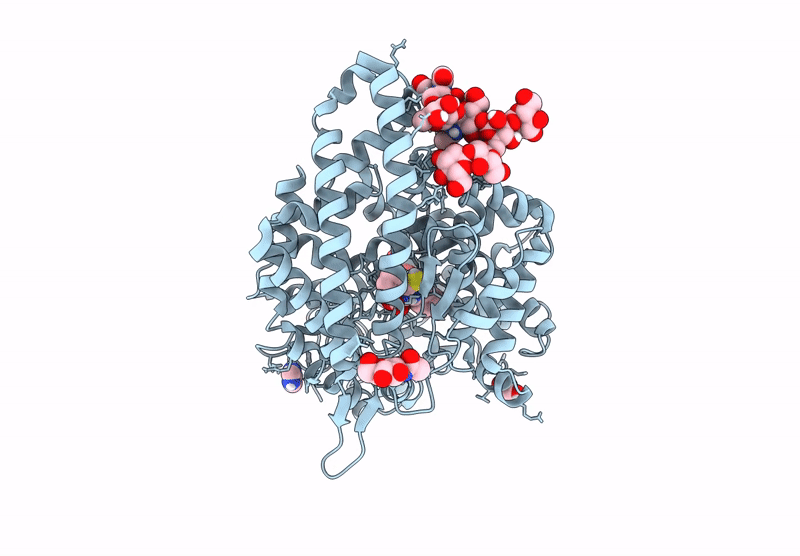
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad014
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-16 Classification: HYDROLASE Ligands: A1IRQ, BO3, PGE, IMD, EDO, NAG, ZN, CL |
 |
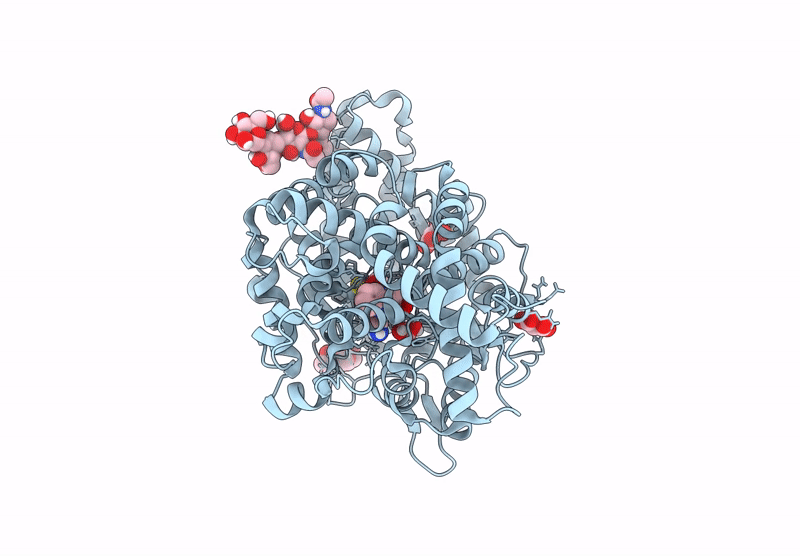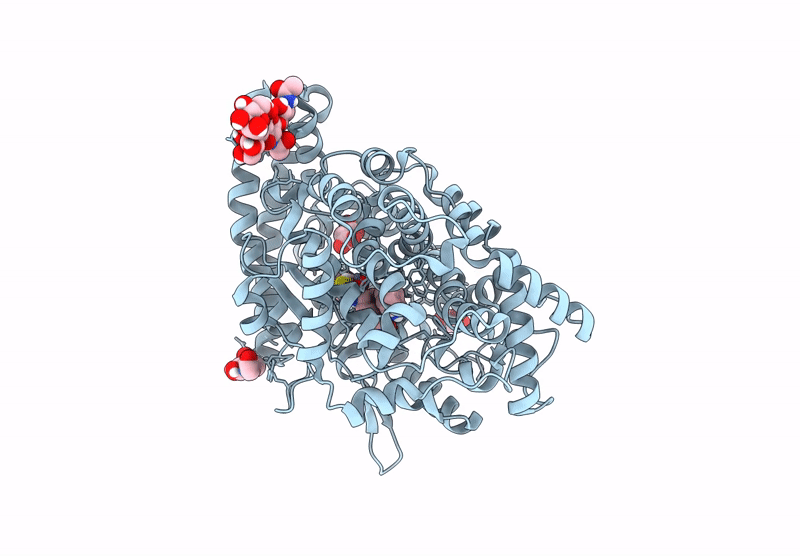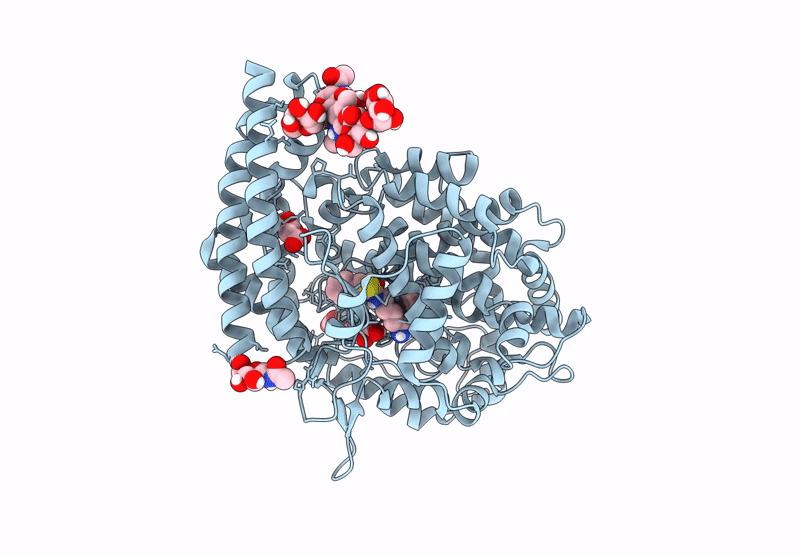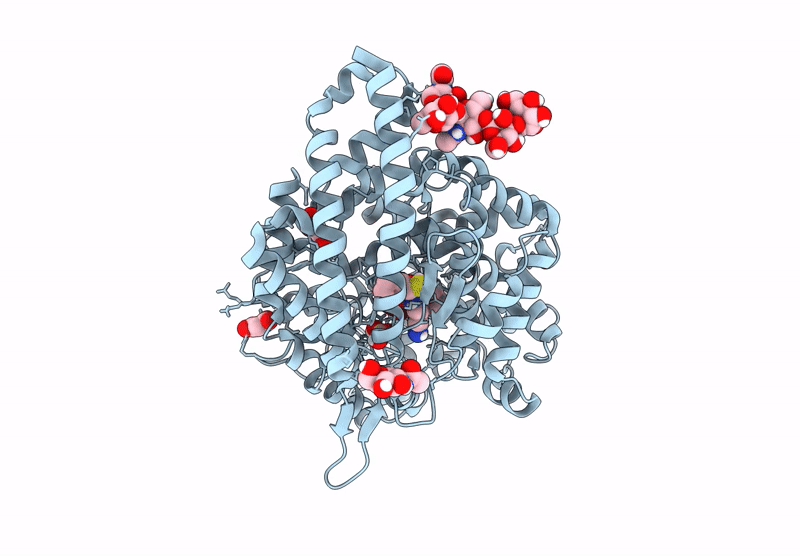
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad015
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-16 Classification: HYDROLASE Ligands: A1IRR, IMD, EDO, NAG, ZN, CL |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad016
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-04-16 Classification: HYDROLASE Ligands: NAG, BO3, MLA, ZN, CL, A1IRS |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme N-Domain In Complex With Dual Ace/Nep Inhibitor Ad011
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: PEG, P33, ACT, EDO, P6G, ZN, CL, MG, 8KC, PG4, NAG |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme N-Domain In Complex With Dual Ace/Nep Inhibitor Ad012
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: 8J9, NAG, ACT, 1PE, EDO, PEG, P33, ZN, CL, MG, PG4 |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme N-Domain In Complex With Dual Ace/Nep Inhibitor Ad013
|
 |
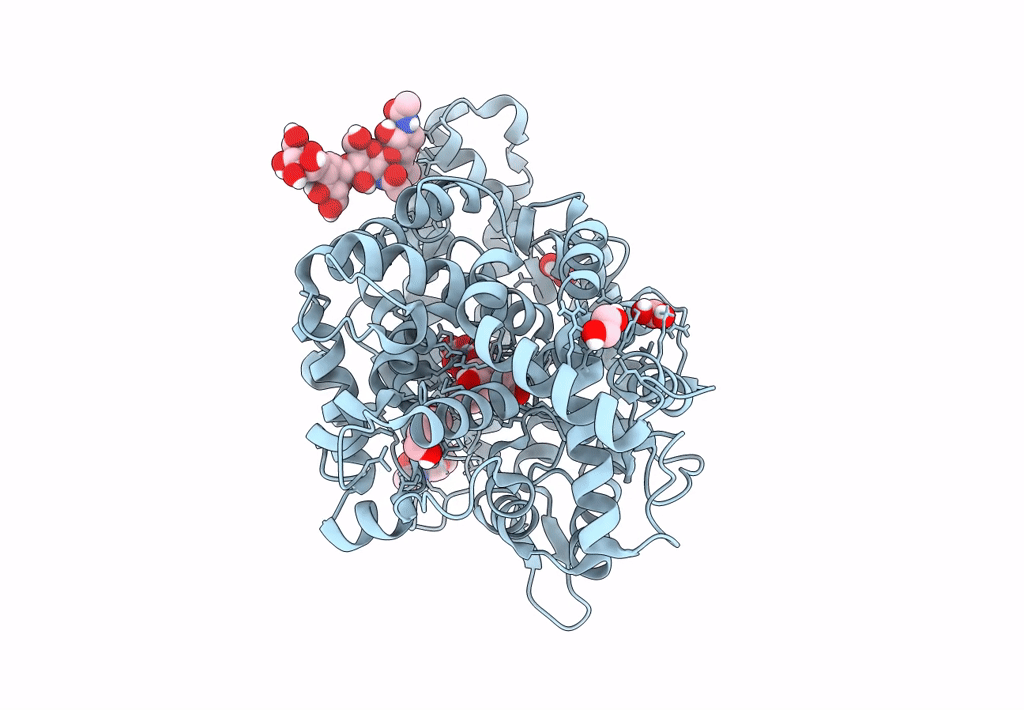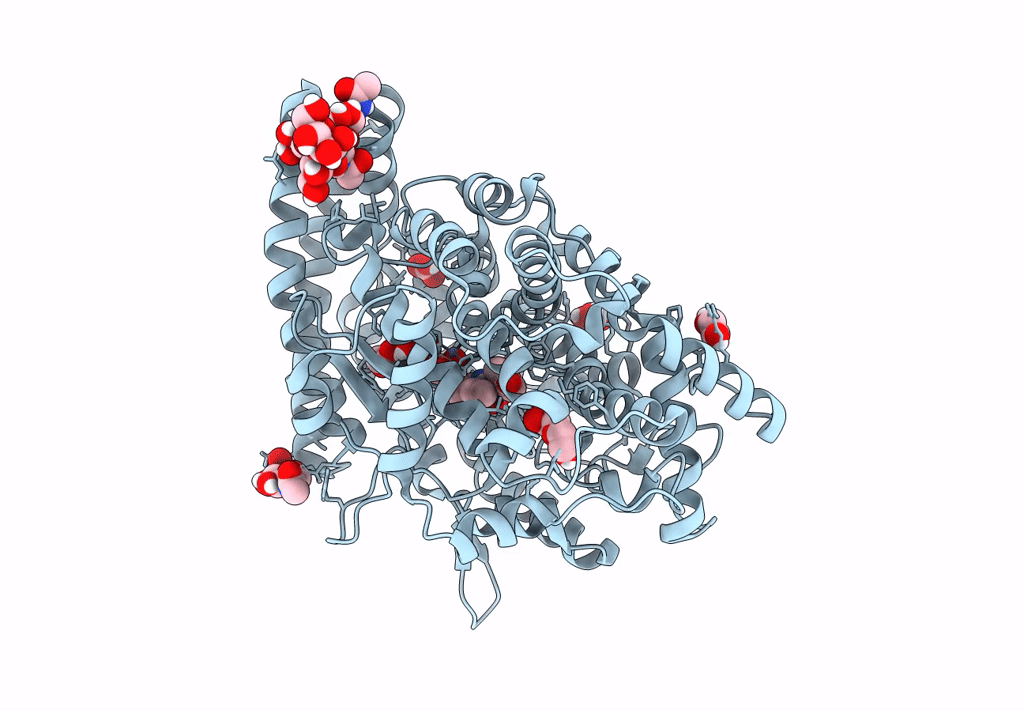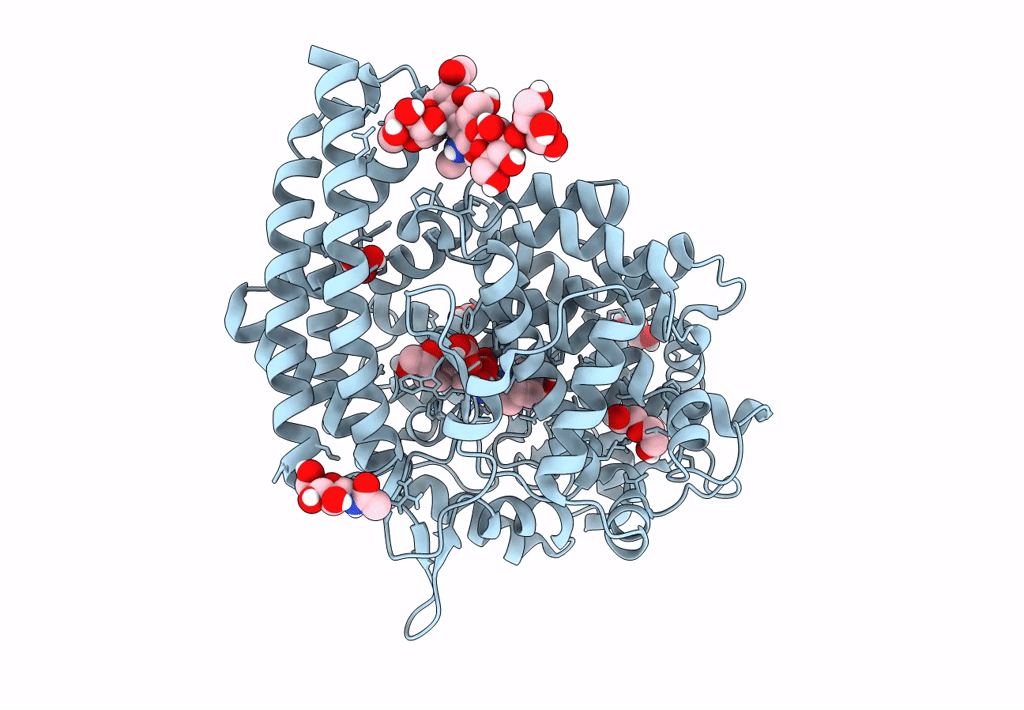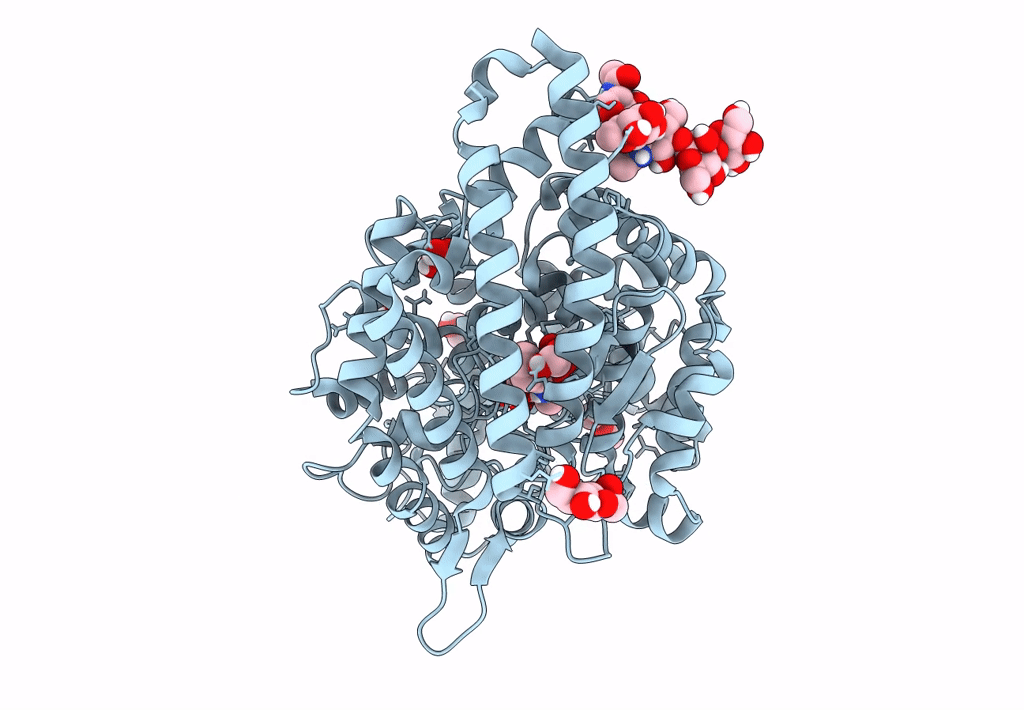
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad011
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: NAG, EDO, ACT, ZN, 8KC, CL, BO3, 1PE, IMD |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad012
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: NAG, BO3, 8J9, PEG, EDO, ZN, CL |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad013
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: NAG, BO3, IMD, 8JV, PEG, 1PE, EDO, ZN, CL |
 |
Organism: Streptococcus pneumoniae ga47502
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-05 Classification: ANTIMICROBIAL PROTEIN Ligands: 54B |
 |
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2014-11-05 Classification: Ribosome/Antibiotic Ligands: MG, ZN, NEG |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-11-13 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 1YM |
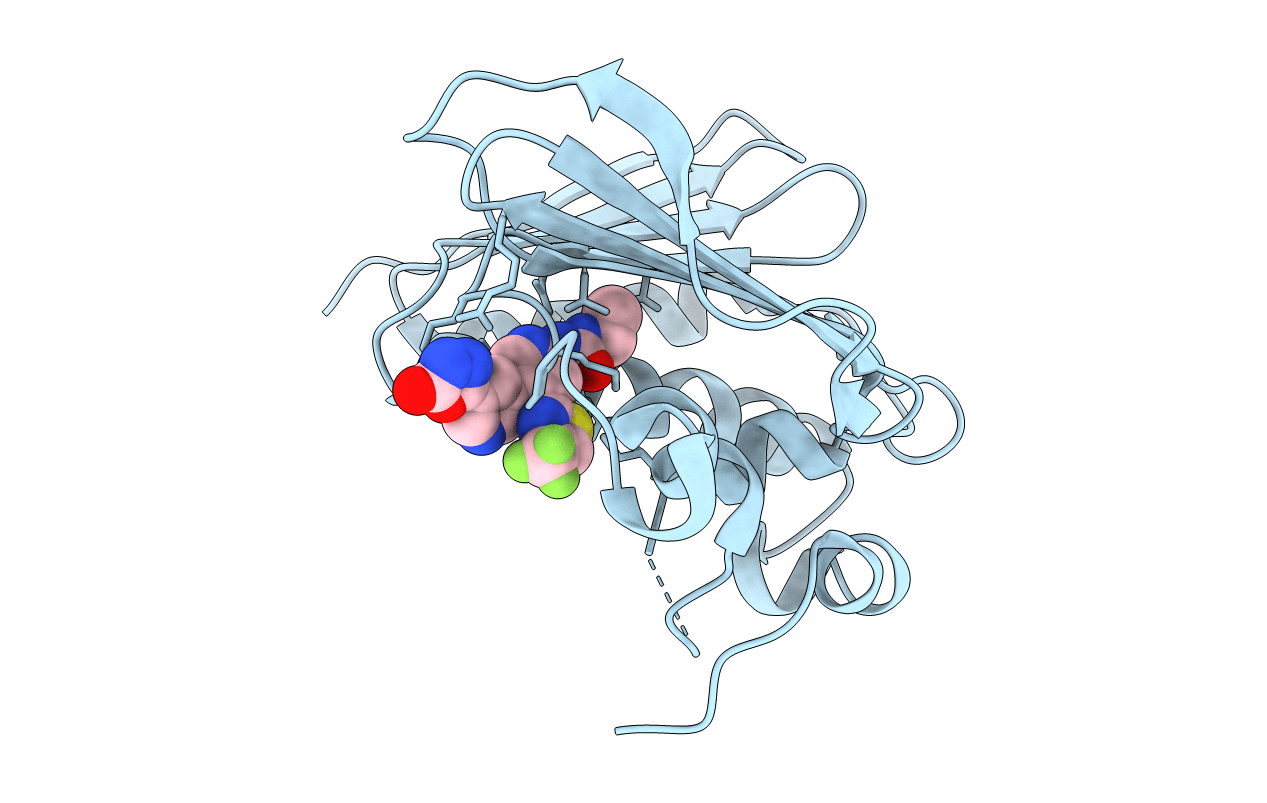 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-10-30 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 1YP |
 |
Design Of Inhibitors Of Helicobacter Pylori Glutamate Racemase As Selective Antibacterial Agents: Incorporation Of Imidazoles Onto A Core Pyrazolopyrimidinedione Scaffold To Improve Bioavailabilty
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-07-18 Classification: ISOMERASE Ligands: KRH, DGL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2011-11-16 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 07N, MG |
 |
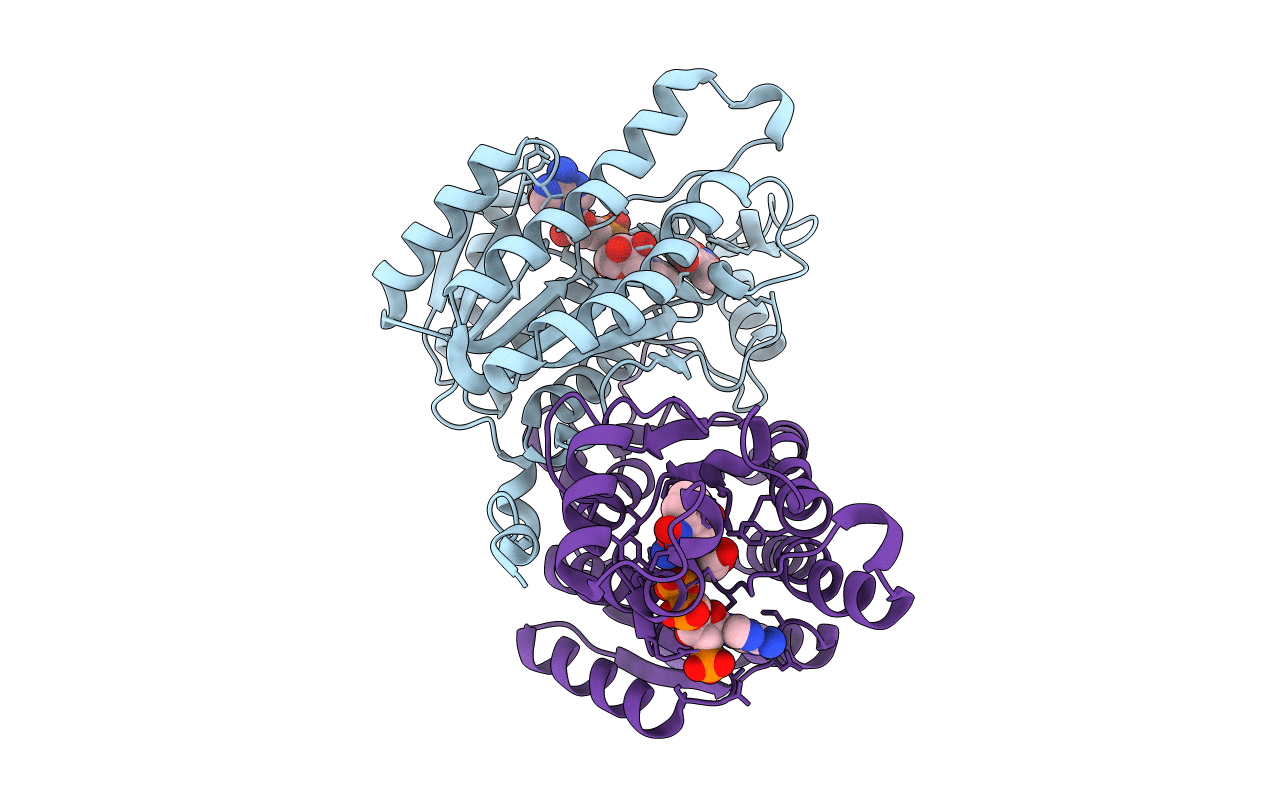
Crystal Structure Of Helicobacter Pylori Glutamate Racemase In Complex With D-Glutamate And An Inhibitor
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2008-12-09 Classification: ISOMERASE Ligands: DGL, VGA |
 |
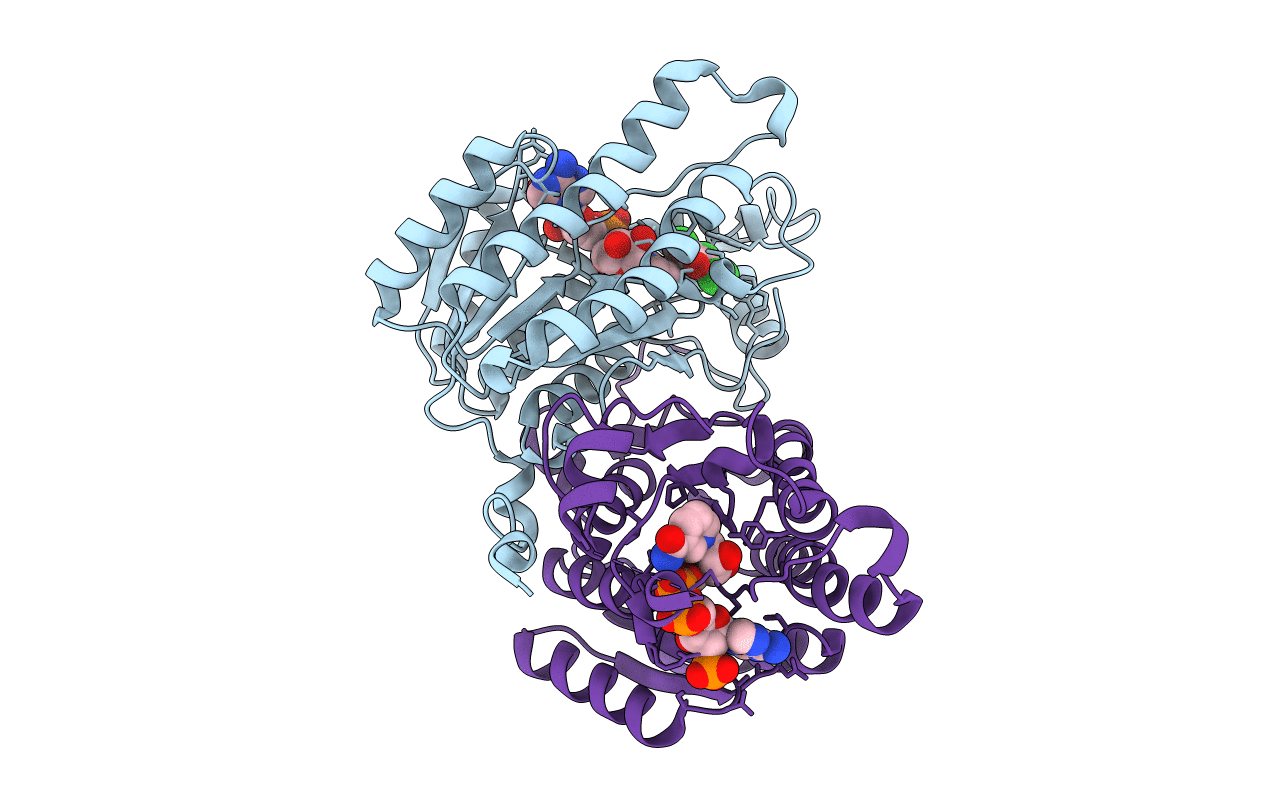
Structure Of Trihydroxynaphthalene Reductase In Complex With Nadph And 4-Nitro-Inden-1-One
Organism: Magnaporthe grisea
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2001-06-06 Classification: OXIDOREDUCTASE Ligands: NDP, NID |
 |
Structure Of Trihydroxynaphthalene Reductase In Complex With Nadph And 4,5,6,7-Tetrachloro-Phthalide
Organism: Magnaporthe grisea
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-06-06 Classification: OXIDOREDUCTASE Ligands: NDP, PHH |

