Search Count: 18
 |
Crystal Structure Of The N-Terminal Kinase Domain From Saccharomyces Cerevisiae Vip1 In Complex With Adp.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, EDO |
 |
Crystal Structure Of The C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, ZN, LPC, ACT |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 In Complex With 1,5-Insp8 (Phosphatase Dead Mutant, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, SPM, ORN, PUT, EDO, I8P, 1JW |
 |
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE |
 |
Alcohol Dehydrogenase From Candida Magnoliae Dsmz 70638 (Adha): Complex With Nadp+
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE Ligands: NAP, IPA, GOL |
 |
Alcohol Dehydrogenase From Candida Magnoliae Dsmz 70638 (Adha): Thermostable 10Fold Mutant
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE |
 |
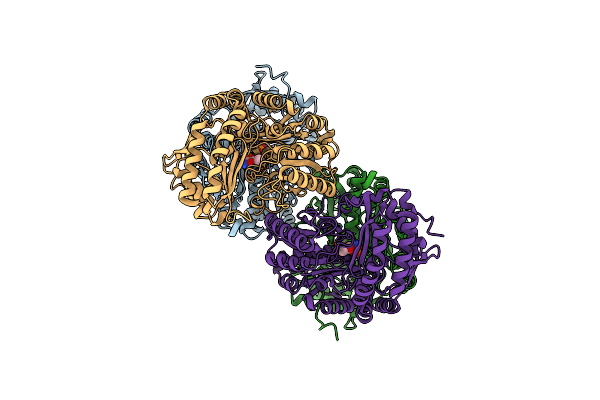
Amine Transaminase Crystal Structure From An Uncultivated Pseudomonas Species In The Plp-Bound (Internal Aldimine) Form
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: PLP |
 |
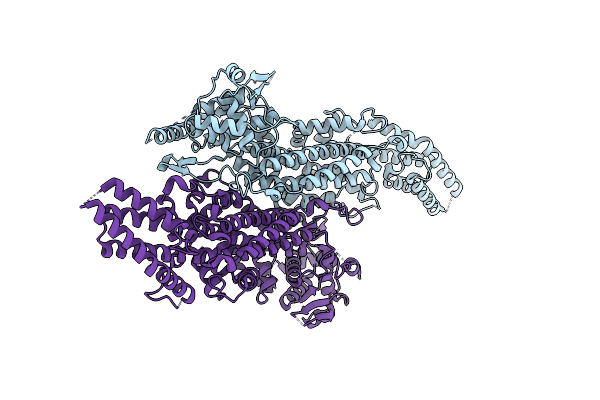
Amine Transaminase Crystal Structure From An Uncultivated Pseudomonas Species In The Pmp-Bound Form
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: PMP |
 |
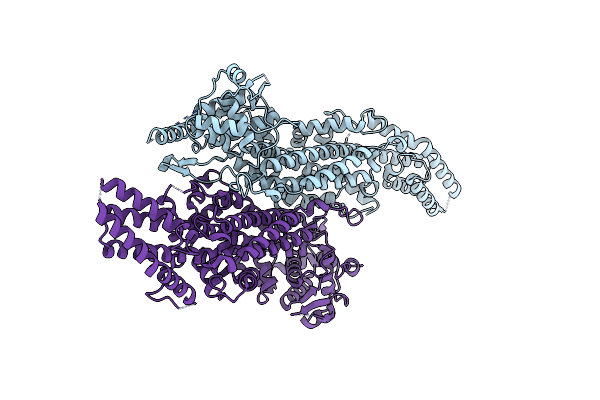
Ironing Out Their Differences: Dissecting The Structural Determinants Of A Phenylalanine Aminomutase And Ammonia Lyase
Organism: Taxus wallichiana var. chinensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-12-10 Classification: LYASE |
 |
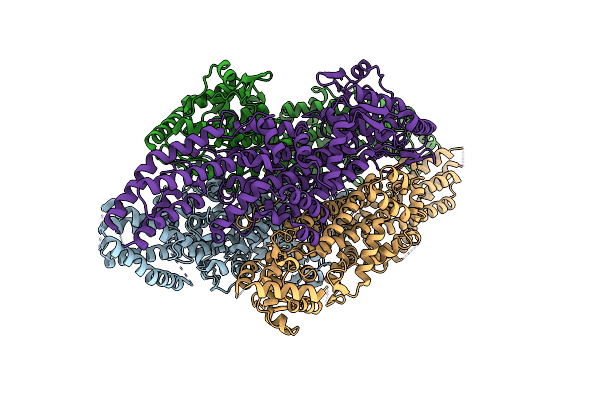
Ironing Out Their Differences: Dissecting The Structural Determinants Of A Phenylalanine Aminomutase And Ammonia Lyase
Organism: Taxus wallichiana var. chinensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-12-10 Classification: LYASE |
 |
Redesign Of A Phenylalanine Aminomutase Into A Beta-Phenylalanine Ammonia Lyase
Organism: Taxus wallichiana var. chinensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-01-16 Classification: LYASE |
 |
Redesign Of A Phenylalanine Aminomutase Into A Beta-Phenylalanine Ammonia Lyase
Organism: Taxus wallichiana var. chinensis
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2013-01-16 Classification: LYASE |
 |
Biochemical Properties And Crystal Structure Of A Novel Beta- Phenylalanine Aminotransferase From Variovorax Paradoxus
Organism: Variovorax paradoxus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2012-10-24 Classification: TRANSFERASE Ligands: IK2, GOL |
 |
Manipulating The Regioselectivity Of Phenylalanine Aminomutase: New Insights Into The Reaction Mechanism Of Mio-Dependent Enzymes From Structure-Guided Directed Evolution
Organism: Taxus wallichiana var. chinensis
Method: X-RAY DIFFRACTION Release Date: 2011-11-09 Classification: LYASE Ligands: BME, FMT, GOL |
 |
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-17 Classification: HYDROLASE |
 |
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-05-18 Classification: HYDROLASE Ligands: SO4 |
 |
Human Neutrophil Gelatinase-Associated Lipocalin (Hngal), Regularised Average Nmr Structure
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1999-05-26 Classification: TRANSPORT PROTEIN |

