Search Count: 17
 |
Chaetomium Thermophilum Mre11-Rad50-Nbs1 Complex Bound To Atpys (Composite Structure)
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: HYDROLASE Ligands: MN, MG, AGS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: HYDROLASE Ligands: MN |
 |
Organism: Thermochaetoides thermophila
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: ZN |
 |
Endonuclease State Of The E. Coli Mre11-Rad50 (Sbccd) Head Complex Bound To Adp And Dsdna
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, MN |
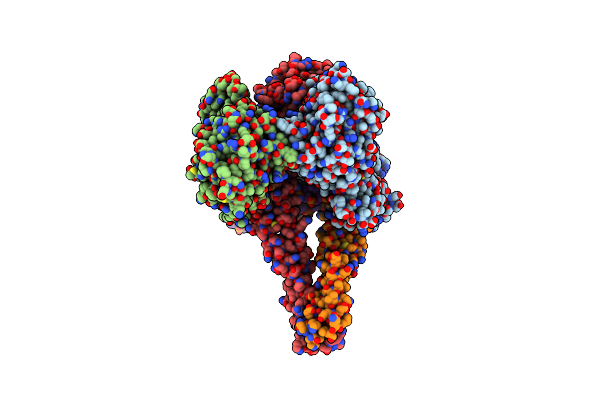 |
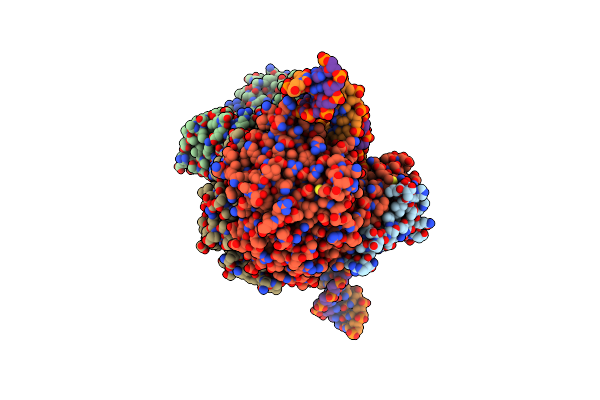
Hairpin-Bound State Of The E. Coli Mre11-Rad50 (Sbccd) Head Complex Bound To Adp And A Dna Hairpin
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: DNA BINDING PROTEIN Ligands: MN, ADP, MG |
 |
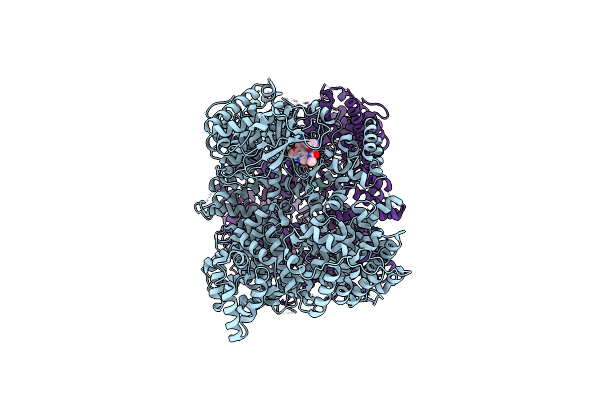
Endonuclease State Of The E. Coli Mre11-Rad50 (Sbccd) Head Complex Bound To Adp And Extended Dsdna
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, MN |
 |
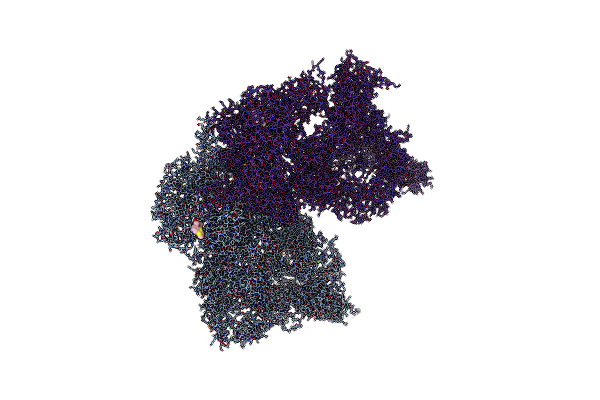
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: SIGNALING PROTEIN Ligands: ZN, UGK |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: SIGNALING PROTEIN Ligands: UF8, ZN |
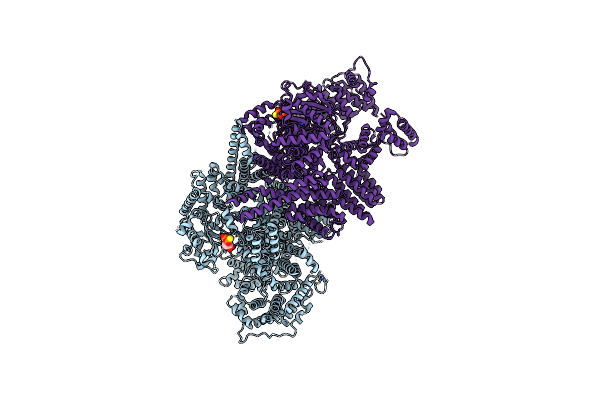 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: SIGNALING PROTEIN Ligands: ZN, AGS, MG |
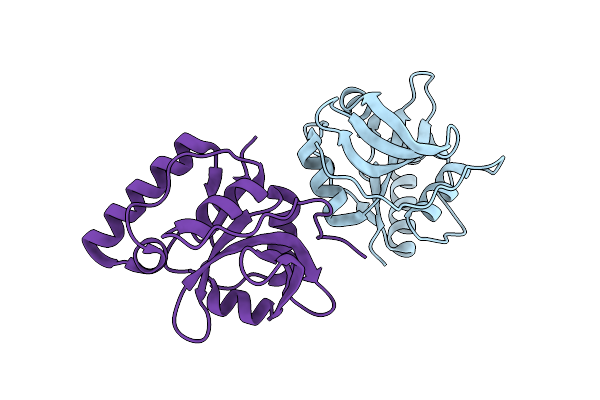 |
Crystal Structure Of The Type Iii Effector Protein Avrrpt2 From Erwinia Amylovora, A C70 Family Cysteine Protease
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-10 Classification: HYDROLASE |
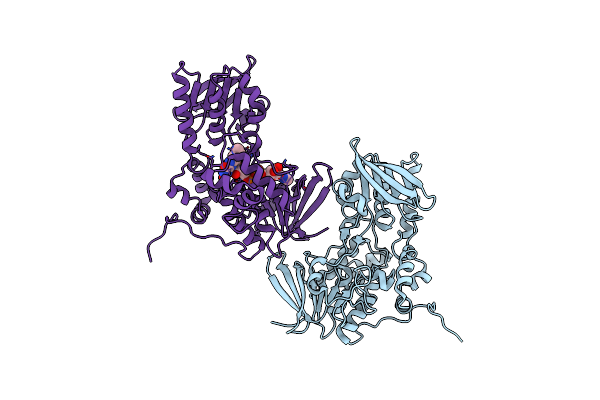 |
The Crystal Structure Of Dfoa Bound To Fad, The Desferrioxamine Biosynthetic Pathway Cadaverine Monooxygenase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-06-27 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD |
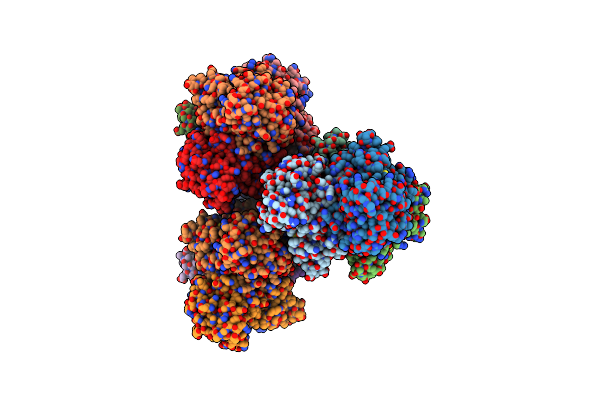 |
Crystal Structure Of Sorbitol-6-Phosphate 2-Dehydrogenase Srld From Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-06-13 Classification: OXIDOREDUCTASE Ligands: CL |
 |
The Crystal Structure Of Dfoj, The Desferrioxamine Biosynthetic Pathway Lysine Decarboxylase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora (strain cfbp1430)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-28 Classification: LYASE Ligands: PLP |
 |
The Crystal Structure Of Dfoc, The Desferrioxamine Biosynthetic Pathway Acetyltransferase/Non-Ribosomal Peptide Synthetase (Nrps)-Independent Siderophore (Nis) From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-02-28 Classification: BIOSYNTHETIC PROTEIN |
 |
The Crystal Structure Of Dfoa Bound To Fad And Nadp; The Desferrioxamine Biosynthetic Pathway Cadaverine Monooxygenase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, NAP |
 |
Semet Crystal Structure Of Erwinia Amylovora Amyr Amylovoran Repressor, A Member Of The Ybjn Protein Family
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2017-02-15 Classification: TRANSCRIPTION |
 |
Erwinia Amylovora Amyr Amylovoran Repressor, A Member Of The Ybjn Protein Family
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-01-18 Classification: SIGNALING PROTEIN |

