Search Count: 22
 |
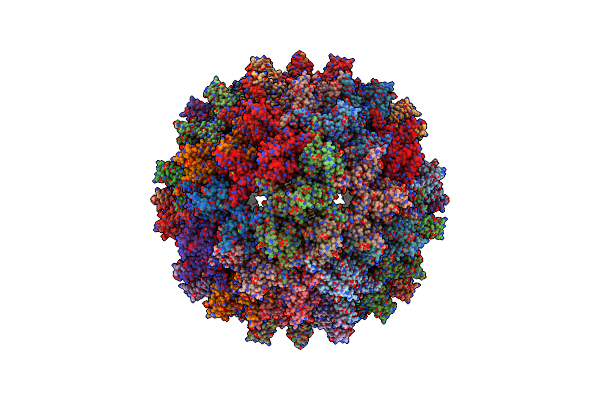
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: A1H7W |
 |
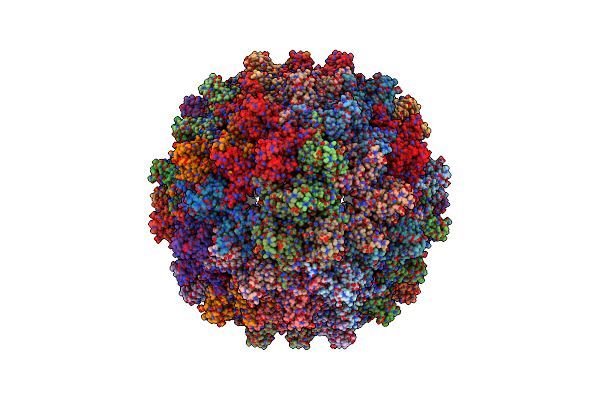
Organism: African cichlid nackednavirus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN |
 |
Organism: African cichlid nackednavirus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN |
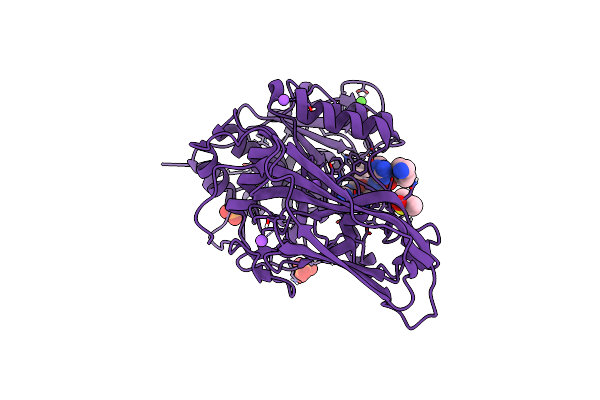 |
X-Ray Structure Of Furin In Complex With The Canavanine-Based Inhibitor 4-Aminomethyl-Phenylacetyl-Canavanine-Tle-Arg-Amba
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: HYDROLASE Ligands: CA, NA, CL, PO4, DMS |
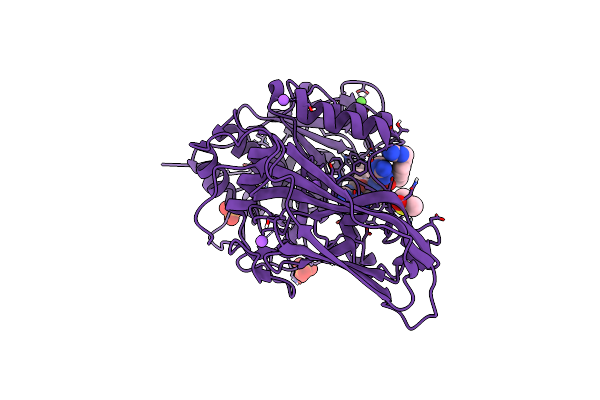 |
X-Ray Structure Of Furin In Complex With The Canavanine Derived Inhibitor 4-Guanidinomethyl-Phenylacetyl-Canavanine-Tle-Arg-Amba
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-17 Classification: HYDROLASE Ligands: CA, NA, CL, PO4, DMS |
 |
X-Ray Structure Of Furin In Complex With The Canavanine-Based Inhibitor 4-Guanidinomethyl-Phenylacetyl-Canavanine-Tle-Canavanine-Amba
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-17 Classification: HYDROLASE Ligands: CA, NA, CL, PO4, DMS |
 |
X-Ray Structure Of Furin In Complex With The Canavanine-Based Inhibitor 4-Guanidinomethyl-Phenylacetyl-Arg-Tle-Canavanine-Amba
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: HYDROLASE Ligands: CA, NA, CL, PO4, DMS |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Proteins On Intact Virions: 3 Closed Rbds
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Hepacivirus c
Method: SOLUTION NMR Release Date: 2019-07-24 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of West Nile Virus Ns2B-Ns3 Protease In Complex With A Capped Dipeptide Boronate Inhibitor
Organism: West nile virus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-12-14 Classification: VIRAL PROTEIN Ligands: 6A8, DMS, GOL |
 |
Organism: Hepatitis c virus jfh-1
Method: SOLUTION NMR Release Date: 2013-06-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Hepatitis c virus
Method: SOLUTION NMR Release Date: 2011-03-02 Classification: VIRAL PROTEIN |
 |
Organism: Hepatitis c virus
Method: SOLUTION NMR Release Date: 2011-03-02 Classification: VIRAL PROTEIN |
 |
Organism: Hepatitis c virus isolate hc-j6
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-01-12 Classification: TRANSFERASE Ligands: 15P, PEG |
 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2011-01-12 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2011-01-12 Classification: TRANSFERASE Ligands: PO4 |
 |
 |
Nmr Structure Of The Membrane Anchor Domain (1-31) Of The Nonstructural Protein 5A (Ns5A) Of Hepatitis C Virus (Minimized Average Structure, Sample In 50% Tfe)
|
 |
Nmr Structure Of The Membrane Anchor Domain (1-31) Of The Nonstructural Protein 5A (Ns5A) Of Hepatitis C Virus (Ensemble Of 51 Structures, Sample In 50% Tfe)
|
 |
Nmr Structure Of The Membrane Anchor Domain (1-31) Of The Nonstructural Protein 5A (Ns5A) Of Hepatitis C Virus (Minimized Average Structure. Sample In 100Mm Sds).
|

