Search Count: 30
 |
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2022-05-11 Classification: VIRAL PROTEIN |
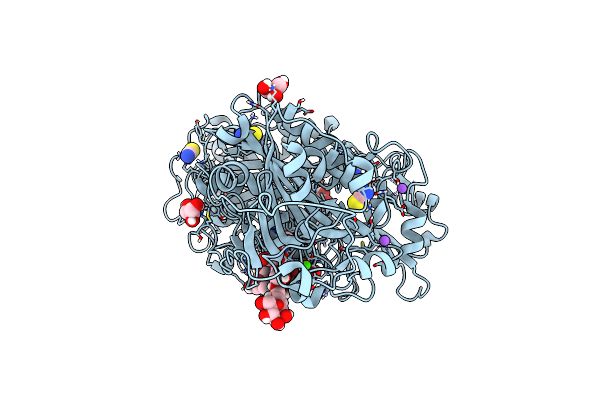 |
Structure-Based Design Of A Novel Class Of Autotaxin Inhibitors Based On Endogenous Allosteric Modulators
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: MAN, ZN, CA, IOD, SCN, NA, GOL, I8K |
 |
Hypoxia-Inducible Factor (Hif) Prolyl Hydroxylase 2 (Phd2) In Complex With The Carboxamide Analog Jnj43058171
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-04-24 Classification: OXIDOREDUCTASE Ligands: FE2, KU1 |
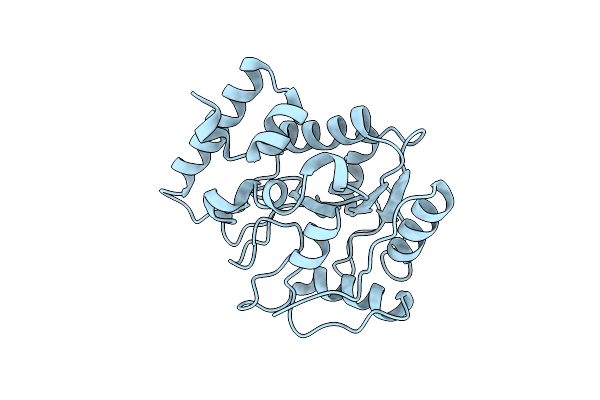 |
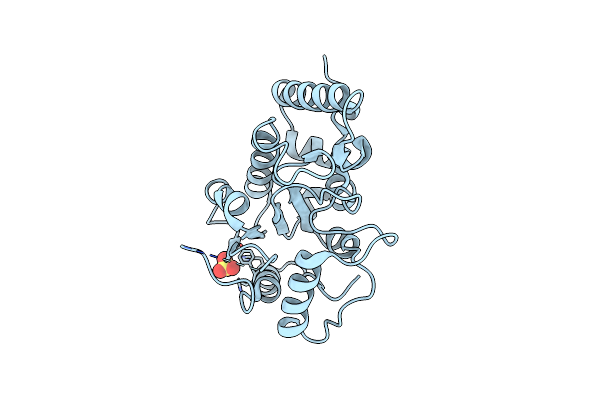
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-03-21 Classification: HYDROLASE |
 |
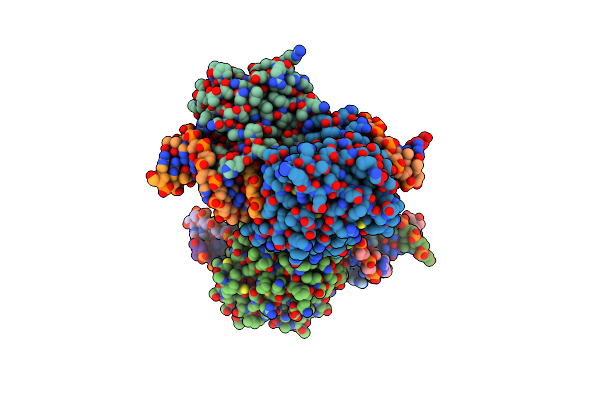
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: SO4 |
 |
Kshv Uracil-Dna Glycosylase, Product Complex With Dsdna Exhibiting Duplex Nucleotide Flipping
Organism: Human herpesvirus 8, synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2018-03-21 Classification: HYDROLASE |
 |
Organism: Streptococcus sp. group g, Human herpesvirus 8, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2017-10-04 Classification: VIRAL PROTEIN |
 |
Organism: Human herpesvirus 8 type m, Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2012-07-25 Classification: HYDROLASE Ligands: ADP |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2012-07-04 Classification: HYDROLASE/DNA Ligands: ADP |
 |
The Crystal Structure Of The Nbd1 Domain Of The Mouse Cftr Protein, Deltaf508 Mutant
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2012-02-01 Classification: HYDROLASE Ligands: ATP, MG, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2011-12-14 Classification: HYDROLASE INHIBITOR |
 |
Organism: Human herpesvirus 8 type m
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-09-21 Classification: DNA BINDING PROTEIN/DNA Ligands: MG, FMT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: FE2, SO4, 014 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: ACT, FE2, 42Z, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: SO4, FE2, AKG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-31 Classification: Hydrolase inhibitor |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2009-03-31 Classification: Hydrolase inhibitor |
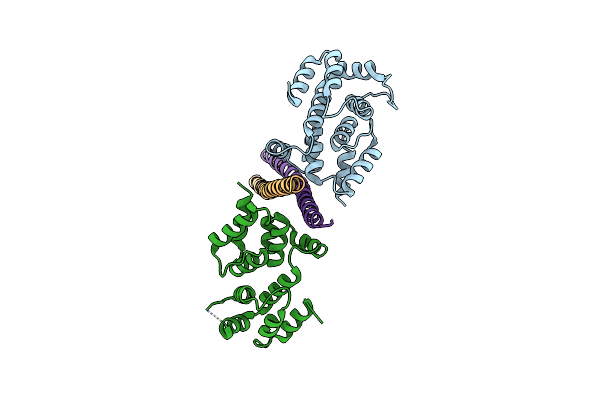 |
Crystal Structure Of A Vflip-Ikkgamma Complex: Insights Into Viral Activation Of The Ikk Signalosome
Organism: Human gammaherpesvirus 8, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2008-06-17 Classification: viral protein/signaling protein |
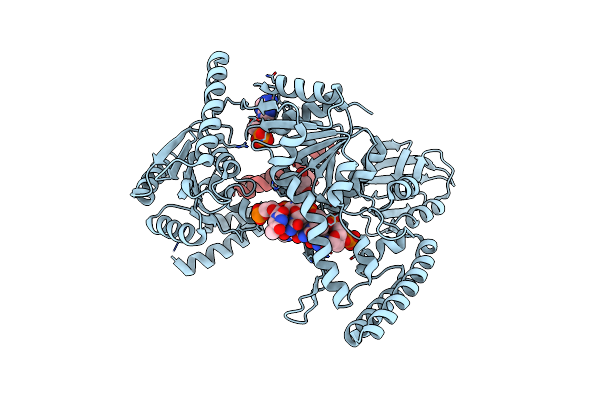 |
Damage Detection By The Uvrabc Pathway: Crystal Structure Of Uvrb Bound To Fluorescein-Adducted Dna
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2007-01-16 Classification: HYDROLASE/DNA Ligands: NML, FLU, ADP |

