Search Count: 9
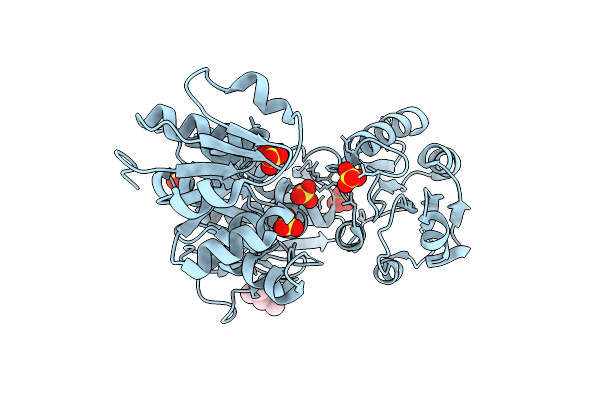 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2016-04-13 Classification: LIGASE Ligands: MPD, NI, SO4 |
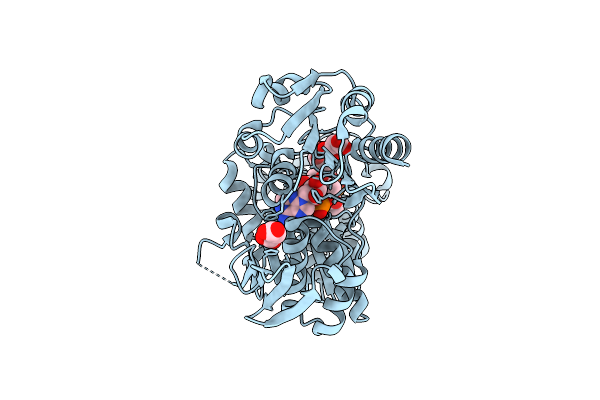 |
Crystal Structure Of Murd Ligase From Escherichia Coli In Complex With Uma And Adp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-13 Classification: LIGASE Ligands: UMA, ADP, MLI |
 |
X-Ray Crystal Structure Of Staphylococcus Aureus Mure With Udp-Murnac- Ala-Glu-Lys And Adp
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-02 Classification: LIGASE Ligands: GOL, UML, ADP, MG |
 |
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2013-07-03 Classification: LIGASE Ligands: IGM |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2013-07-03 Classification: LIGASE Ligands: 2GN |
 |
Structure Of Paem, A Colicin M-Like Bacteriocin Produced By Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-09-12 Classification: HYDROLASE Ligands: MG, EDO |
 |
Structure Of Paem, A Colicin M-Like Bacteriocin Produced By Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2012-09-12 Classification: HYDROLASE |
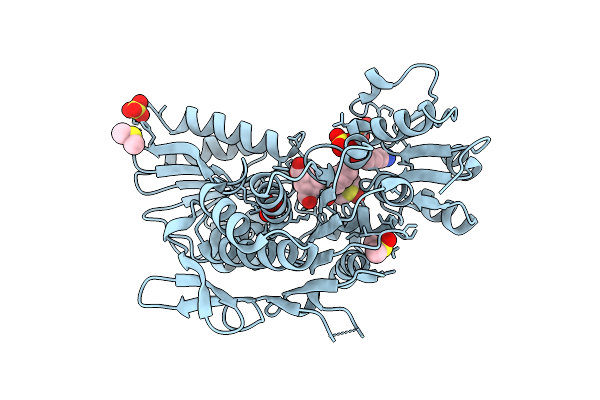 |
Second-Generation Sulfonamide Inhibitors Of Murd: Activity Optimisation With Conformationally Rigid Analogues Of D-Glutamic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2011-05-18 Classification: LIGASE Ligands: 051, DMS, SO4, CL |
 |
The X-Ray Structure Of The Escherichia Coli Colicin M Immunity Protein Demonstrates The Presence Of A Disulphide Bridge, Which Is Functionally Essential
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-11-17 Classification: ANTIBIOTIC Ligands: CD, CL, NA |

