Search Count: 19
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Xchem Fragment Z19735904 At 1.14 Angstrom Resolution.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: B0A, SO4, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#827 At 1.40 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX2, CL, SO4, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#909 At 1.61 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX3, SO4, DMS, CL |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#985 At 1.65 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX4, SO4, DMS, CL |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1004 At 1.40 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IXY, SO4, CL, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1010 At 1.50 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IXZ, SO4, CL, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1032 At 1.61 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX0, SO4, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1090 At 1.74 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX1, SO4, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-02-26 Classification: TRANSFERASE Ligands: SO4, A1IYU, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: TRANSFERASE Ligands: A1IYV, SO4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-26 Classification: TRANSFERASE Ligands: SO4, EDO, A1IYW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-26 Classification: TRANSFERASE Ligands: SO4, A1IYX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-02-26 Classification: TRANSFERASE Ligands: SO4, A1IYY, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-02-26 Classification: TRANSFERASE Ligands: SO4, A1IYZ, EDO |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-09 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, LNU, IMD, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-11-09 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, LO9, ACP |
 |
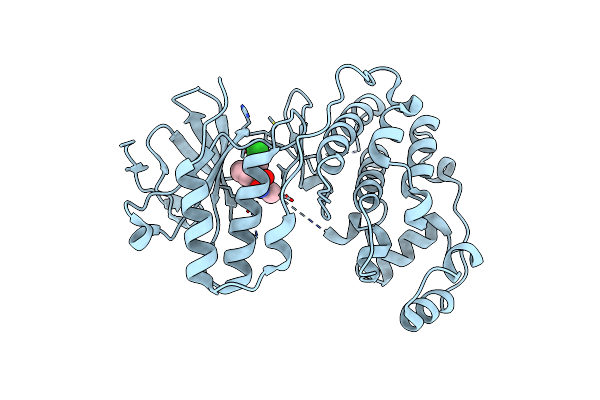
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: BOG, 9Y8 |
 |
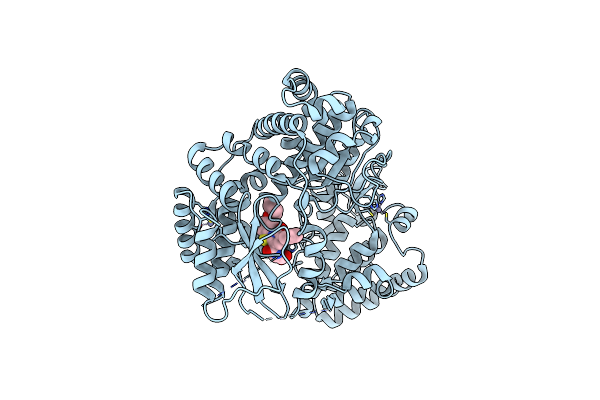
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: 9Y5 |
 |
Crystal Structure Of Dengue Virus Serotype 3 Rna Dependent Rna Polymerase Bound To Hee1-2Tyr, A New Pyridobenzothizole Inhibitor
Organism: Dengue virus 3
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-10-05 Classification: HYDROLASE Ligands: ZN, 6CJ |

