Search Count: 4
 |
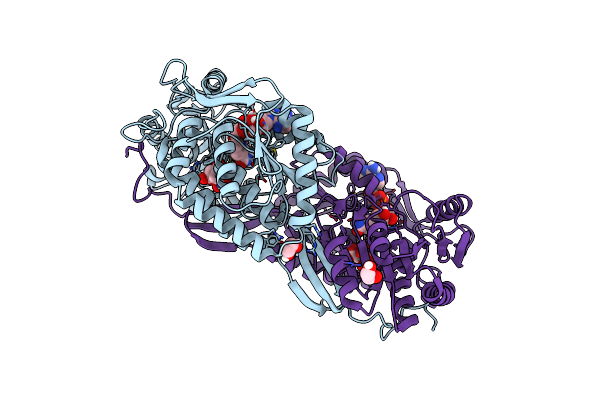
X-Ray Crystal Structure Of Fatty Aldehyde Dehydrogenase Enzymes From Marinobacter Aquaeolei Vt8 Complexed With A Substrate
Organism: Marinobacter hydrocarbonoclasticus (strain atcc 700491 / dsm 11845 / vt8)
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE Ligands: EDO, 8YP, MPD, CL, PO4 |
 |
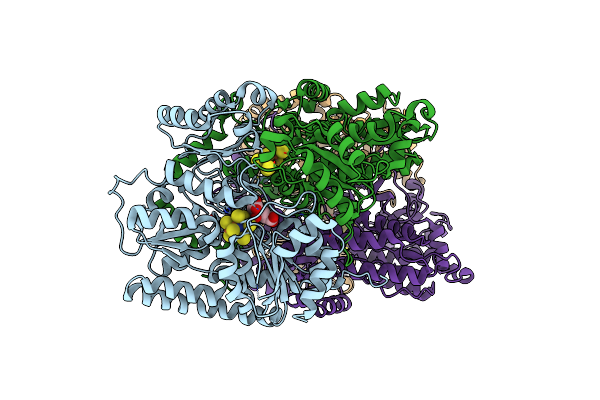
Fatty Aldehyde Dehydrogenase From Marinobacter Aquaeolei Vt8 And Cofactor Complex
Organism: Marinobacter hydrocarbonoclasticus (strain atcc 700491 / dsm 11845 / vt8)
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE Ligands: EDO, CIT, NAD |
 |
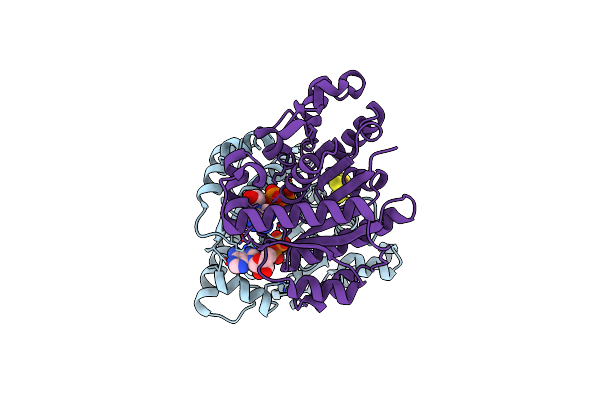
Insights Into Substrate Binding At Femo-Cofactor In Nitrogenase From The Structure Of An Alpha-70Ile Mofe Protein Variant
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2010-02-16 Classification: OXIDOREDUCTASE Ligands: HCA, CFN, CLF, CA |
 |
Crystal Structure Of The L Protein Of Rhodobacter Sphaeroides Light-Independent Protochlorophyllide Reductase (Bchl) With Mgadp Bound: A Homologue Of The Nitrogenase Fe Protein
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE Ligands: MG, ADP, SF4 |

