Search Count: 15
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, WNT |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, DMS, WNC |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, WN8 |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, WMC, DMS |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, DMS, WMN |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, DMS, WMI |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, WN2 |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, DMS, WMX |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, WM6 |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, DMS, WLZ |
 |
Crystal Structure Of Erk2 In Complex With A Covalently Bound Macrocyclic Ligand
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-04-16 Classification: SIGNALING PROTEIN Ligands: SO4, WMT |
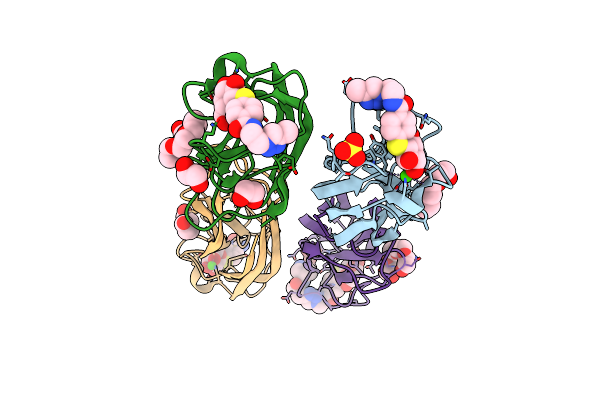 |
Crystal Structure Of Leca Lectin Complexed With A Divalent Galactoside At 1.65 Angstrom
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-10-08 Classification: SUGAR BINDING PROTEIN Ligands: CA, GAL, CN8, 1PE, SO4, EDO |
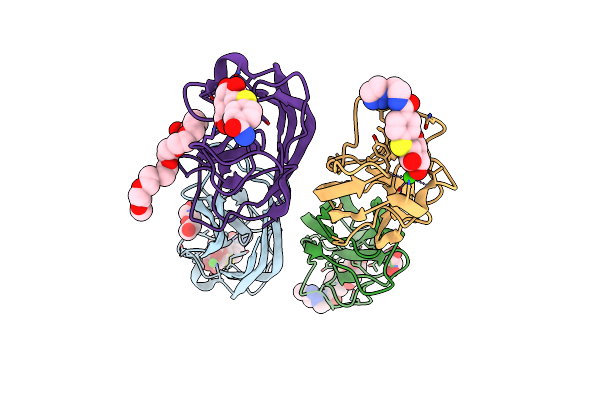 |
Crystal Structure Of Leca In Complex With A Divalent Galactoside At 1. 57 Angstrom In Magnesium
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2014-10-08 Classification: SUGAR BINDING PROTEIN Ligands: CA, GAL, CN8, PEG, EDO, 2PE, CL |
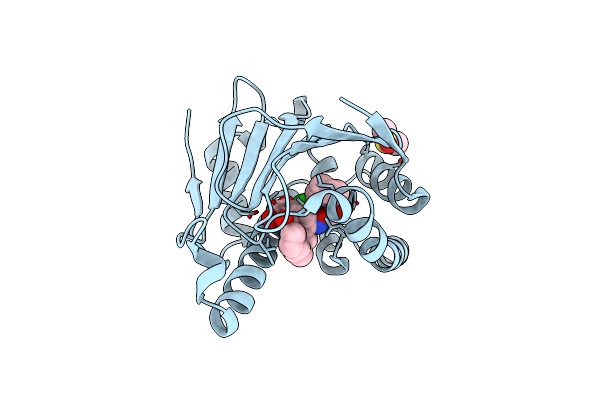 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-08-18 Classification: CHAPERONE Ligands: DMS, JZB |
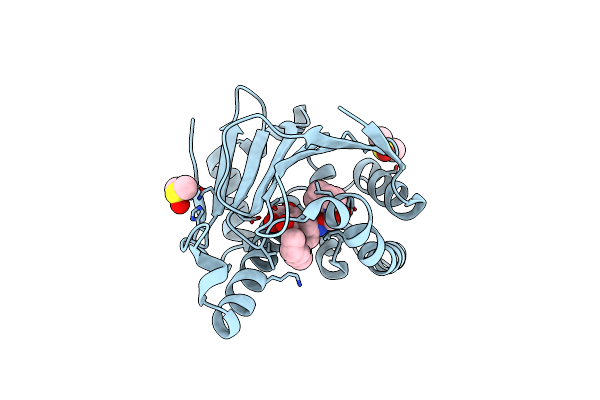 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-08-18 Classification: CHAPERONE Ligands: DMS, JZC |

