Search Count: 15
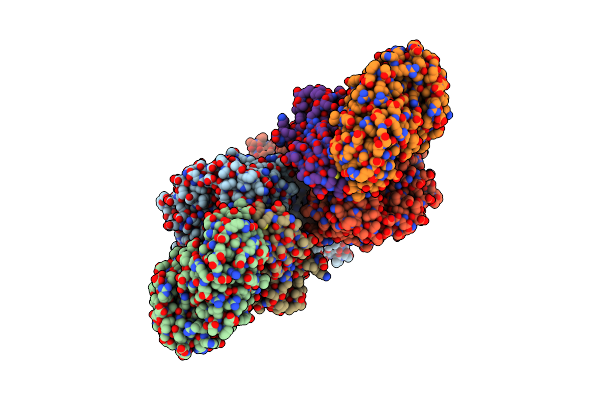 |
The Cryoem Structure Of The High Affinity Carbon Monoxide Dehydrogenase From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: CUN, MCN, FAD, FES |
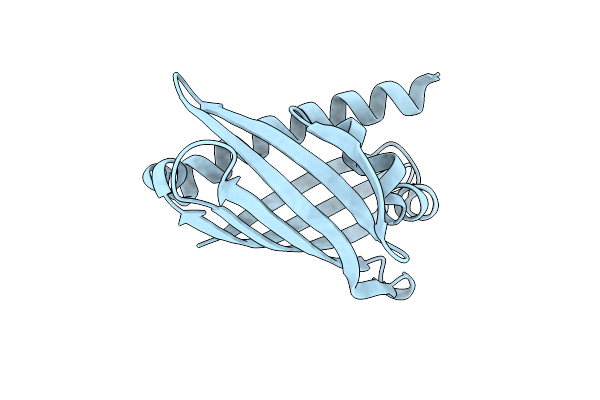 |
The Crystal Structure Of Coxg From M. Smegmatis, Minus Lipid Anchoring C-Terminus.
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: ELECTRON TRANSPORT |
 |
The 2.19-Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Complex Minus Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.67 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, OH, MG, VK3, F3S |
 |
The Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Full Complex Focused Refinement Of Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.52 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: O, MG, FCO, 3NI, VK3, F3S |
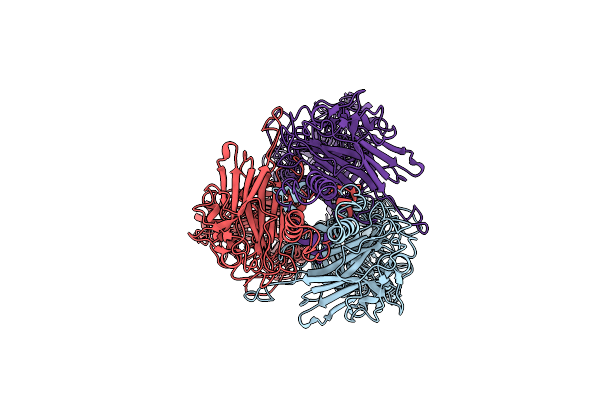 |
Organism: Klebsiella phage gh-k3
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: SUGAR BINDING PROTEIN |
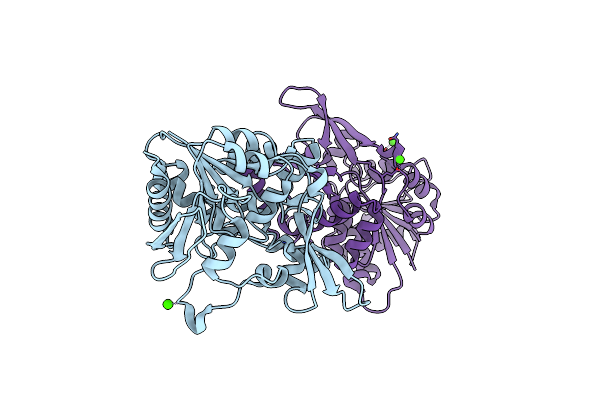 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: CA |
 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FO1, CA |
 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, GOL, CA |
 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Gdp And Fo Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, FO1, CA |
 |
The Crystal Structure Of Fbia From Mycobacterium Smegmatis, Dehydro-F420-0 Bound Form
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, QMD, CA |
 |
Abc Transporter-Associated Periplasmic Binding Protein Dppa From Helicobacter Pylori In Complex With Peptide Stsa
Organism: Helicobacter pylori ss1, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-08-21 Classification: Periplasmic peptide binding protein |
 |
Abc Transporter-Associated Periplasmic Binding Protein Dppa From Helicobacter Pylori
Organism: Helicobacter pylori, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-12-20 Classification: CELL CYCLE/INHIBITOR Ligands: 8QT |

