Search Count: 161
 |
 |
 |
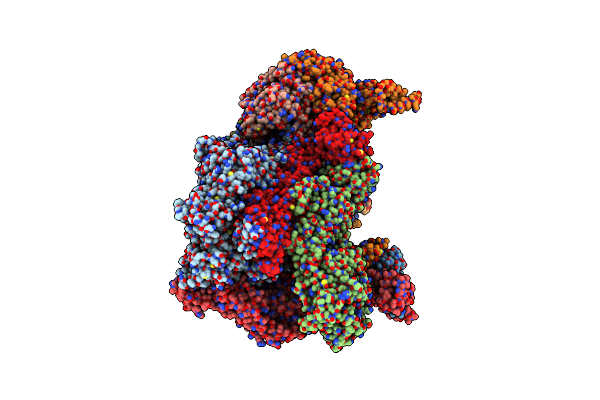 |
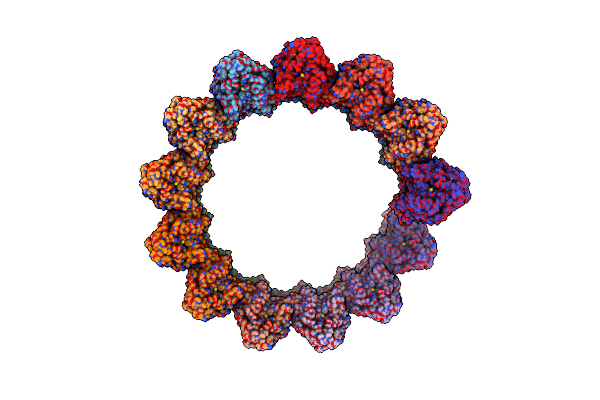
High-Resolution Structure Of The Anaphase-Promoting Complex/Cyclosome (Apc/C) Bound To Co-Activator Cdh1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: CELL CYCLE Ligands: ZN |
 |
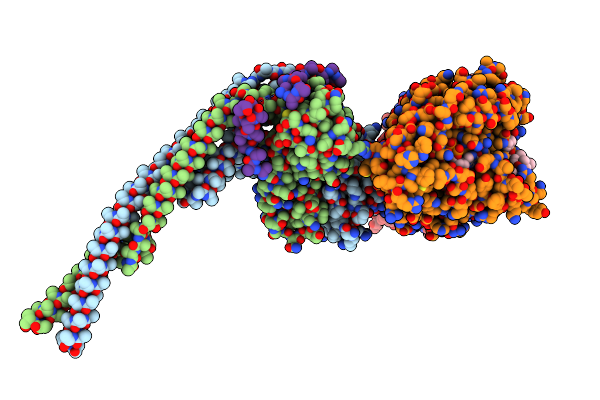
Structure Of The Native Microtubule Lattice Nucleated From The Yeast Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE |
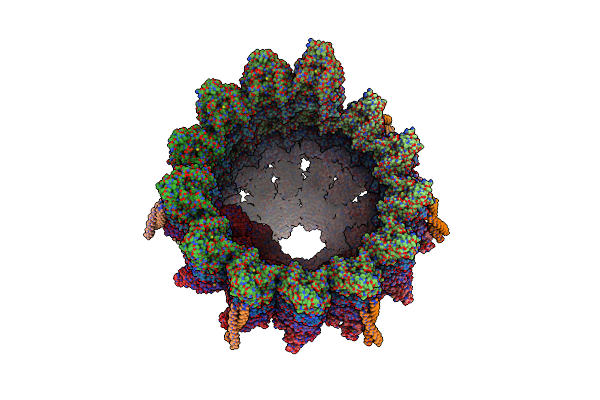 |
Structure Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP, GDP |
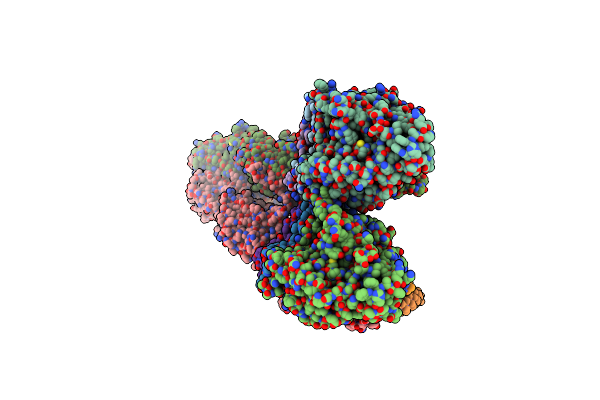 |
Structure Of The Y-Tubulin Small Complex (Ytusc) As Part Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP |
 |
Organism: Saccharomyces cerevisiae, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: CELL CYCLE Ligands: GTP, MG, GDP, TA1 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-04-03 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: CELL CYCLE Ligands: HOH |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
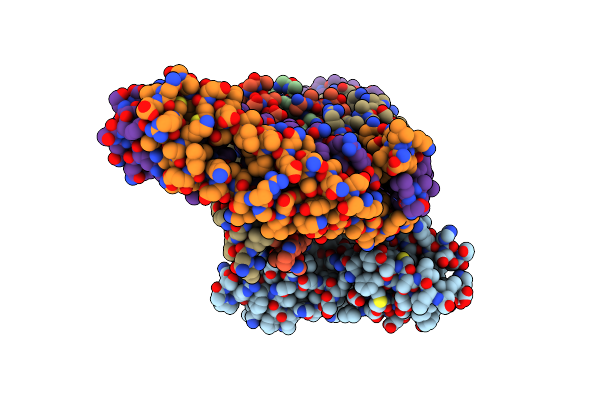 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
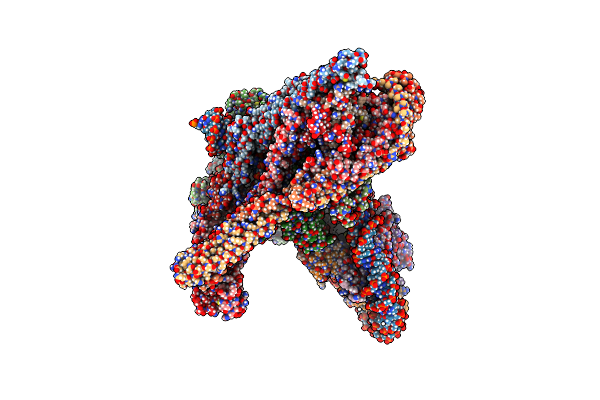 |
Cryo-Em Structure Of Cbf1-Ccan Bound Topologically To A Centromeric Cenp-A Nucleosome
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
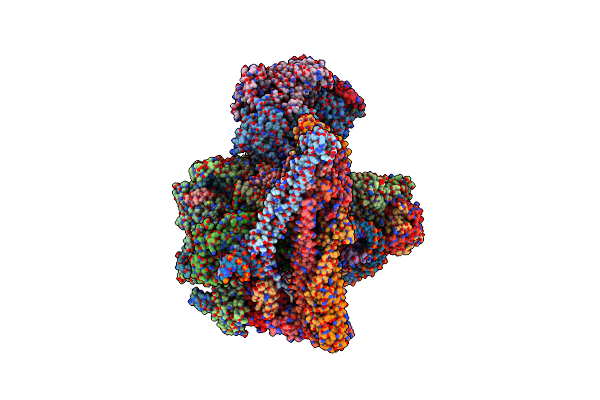 |
Cryo-Em Structure Of The Yeast Inner Kinetochore Bound To A Cenp-A Nucleosome.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
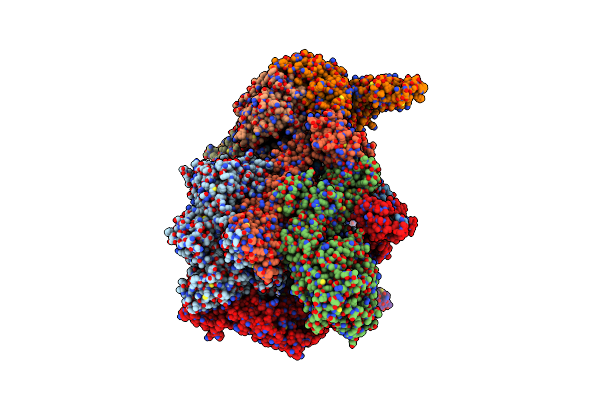 |
Cryo-Em Structure Of The Apo Anaphase-Promoting Complex/Cyclosome (Apc/C) At 3.2 Angstrom Resolution
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: CELL CYCLE Ligands: ZN |

